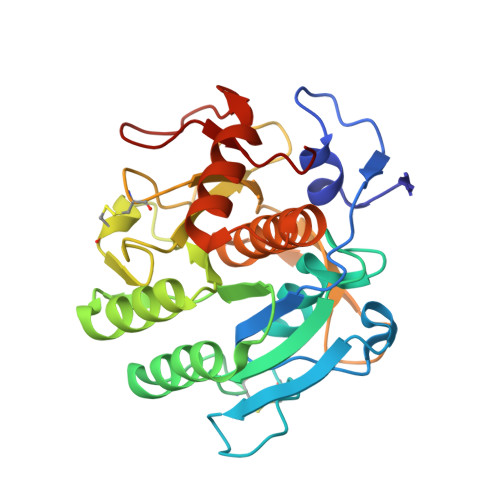
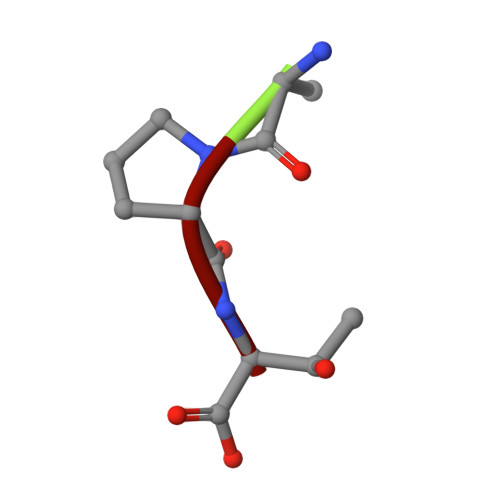
The 1.8 A crystal structure of a proteinase K-like enzyme from a psychrotroph Serratia species
Helland, R., Larsen, A.N., Smalas, A.O., Willassen, N.P.(2006) FEBS J 273: 61-71
- PubMed: 16367748
- DOI: https://doi.org/10.1111/j.1742-4658.2005.05040.x
- Primary Citation of Related Structures:
2B6N - PubMed Abstract:
Proteins from organisms living in extreme conditions are of particular interest because of their potential for being templates for redesign of enzymes both in biotechnological and other industries. The crystal structure of a proteinase K-like enzyme from a psychrotroph Serratia species has been solved to 1.8 A. The structure has been compared with the structures of proteinase K from Tritirachium album Limber and Vibrio sp. PA44 in order to reveal structural explanations for differences in biophysical properties. The Serratia peptidase shares around 40 and 64% identity with the Tritirachium and Vibrio peptidases, respectively. The fold of the three enzymes is essentially identical, with minor exceptions in surface loops. One calcium binding site is found in the Serratia peptidase, in contrast to the Tritirachium and Vibrio peptidases which have two and three, respectively. A disulfide bridge close to the S2 site in the Serratia and Vibrio peptidases, an extensive hydrogen bond network in a tight loop close to the substrate binding site in the Serratia peptidase and different amino acid sequences in the S4 sites are expected to cause different substrate specificity in the three enzymes. The more negative surface potential of the Serratia peptidase, along with a disulfide bridge close to the S2 binding site of a substrate, is also expected to contribute to the overall lower binding affinity observed for the Serratia peptidase. Clear electron density for a tripeptide, probably a proteolysis product, was found in the S' sites of the substrate binding cleft.
- Norwegian Structural Biology Centre, Faculty of Science, University of Tromsø, Tromsø, Norway. ronny.helland@chem.uit.no
Organizational Affiliation: