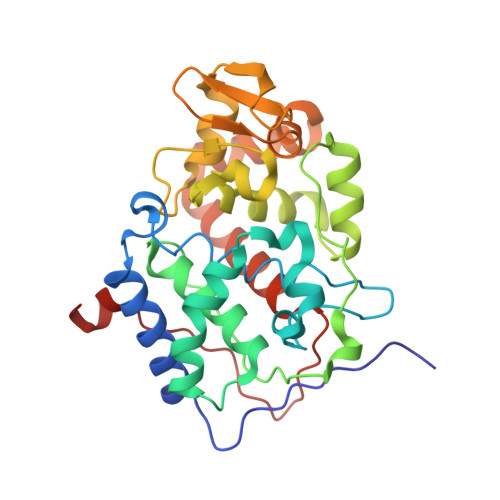
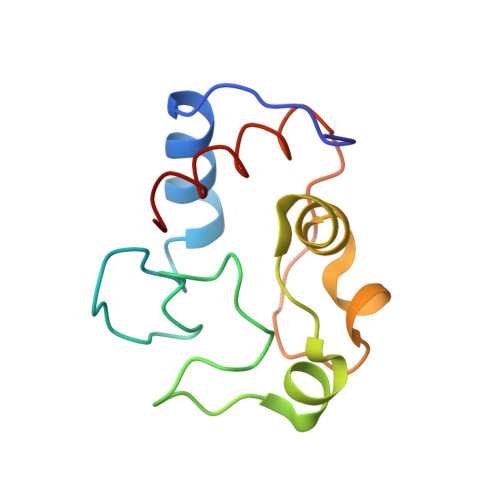
Effects of interface mutations on association modes and electron-transfer rates between proteins
Kang, S.A., Crane, B.R.(2005) Proc Natl Acad Sci U S A 102: 15465-15470
- PubMed: 16227441
- DOI: https://doi.org/10.1073/pnas.0505176102
- Primary Citation of Related Structures:
2B0Z, 2B10, 2B11, 2B12 - PubMed Abstract:
Although bonding networks determine electron-transfer (ET) rates within proteins, the mechanism by which structure and dynamics influence ET across protein interfaces is not well understood. Measurements of photochemically induced ET and subsequent charge recombination between Zn-porphyrin-substituted cytochrome c peroxidase and cytochrome c in single crystals correlate reactivity with defined structures for different association modes of the redox partners. Structures and ET rates in crystals are consistent with tryptophan oxidation mediating charge recombination reactions. Conservative mutations at the interface can drastically affect how the proteins orient and dispose redox centers. Whereas some configurations are ET inactive, the wild-type complex exhibits the fastest recombination rate. Other association modes generate ET rates that do not correlate with predictions based on cofactor separations or simple bonding pathways. Inhibition of photoinduced ET at <273 K indicates gating by small-amplitude dynamics, even within the crystal. Thus, different associations achieve states of similar reactivity, and within those states conformational fluctuations enable interprotein ET.
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA.
Organizational Affiliation: