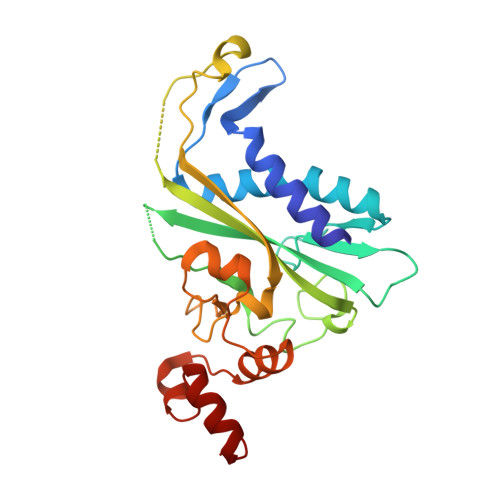
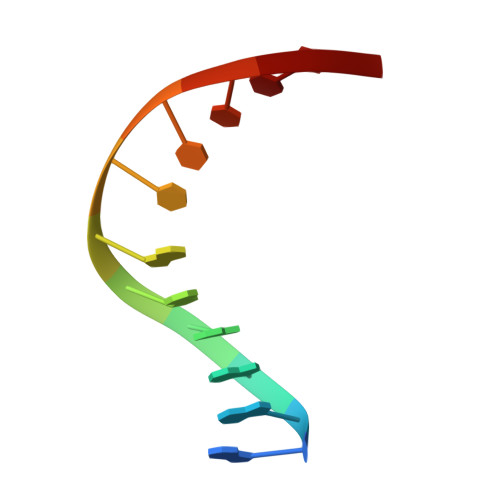
Non-cognate Enzyme-DNA Complex: Structural and Kinetic Analysis of EcoRV Endonuclease Bound to the EcoRI Recognition Site GAATTC
Hiller, D.A., Rodriguez, A.M., Perona, J.J.(2005) J Mol Biology 354: 121-136
- PubMed: 16236314
- DOI: https://doi.org/10.1016/j.jmb.2005.09.046
- Primary Citation of Related Structures:
2B0D, 2B0E - PubMed Abstract:
The crystal structure of EcoRV endonuclease bound to non-cognate DNA at 2.0 angstroms resolution shows that very small structural adaptations are sufficient to ensure the extreme sequence specificity characteristic of restriction enzymes. EcoRV bends its specific GATATC site sharply by 50 degrees into the major groove at the center TA step, generating unusual base-base interactions along each individual DNA strand. In the symmetric non-cognate complex bound to GAATTC, the center step bend is relaxed to avoid steric hindrance caused by the different placement of the exocyclic thymine methyl groups. The decreased base-pair unstacking in turn leads to small conformational rearrangements in the sugar-phosphate backbone, sufficient to destabilize binding of crucial divalent metal ions in the active site. A second crystal structure of EcoRV bound to the base-analog GAAUTC site shows that the 50 degrees center-step bend of the DNA is restored. However, while divalent metals bind at high occupancy in this structure, one metal ion shifts away from binding at the scissile DNA phosphate to a position near the 3'-adjacent phosphate group. This may explain why the 10(4)-fold attenuated cleavage efficiency toward GAATTC is reconstituted by less than tenfold toward GAAUTC. Examination of DNA binding and bending by equilibrium and stopped-flow florescence quenching and fluorescence resonance energy transfer (FRET) methods demonstrates that the capacity of EcoRV to bend the GAATTC non-cognate site is severely limited, but that full bending of GAAUTC is achieved at only a threefold reduced rate compared with the cognate complex. Together, the structural and biochemical data demonstrate the existence of distinct mechanisms for ensuring specificity at the bending and catalytic steps, respectively. The limited conformational rearrangements observed in the EcoRV non-cognate complex provide a sharp contrast to the extensive structural changes found in a non-cognate BamHI-DNA crystal structure, thus demonstrating a diversity of mechanisms by which restriction enzymes are able to achieve specificity.
- Department of Chemistry and Biochemistry, and Interdepartmental Program in Biomolecular Science and Engineering, University of California at Santa Barbara, Santa Barbara, CA 93106-9510, USA.
Organizational Affiliation: