CU/ZN superoxide dismutase from neisseria meningitidis K91E, K94E double mutant
DiDonato, M., Kassmann, C.J., Bruns, C.K., Cabelli, D.E., Cao, Z., Tabatabai, L.B., Kroll, J.S., Getzoff, E.D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Superoxide dismutase [Cu-Zn] | 164 | Neisseria meningitidis | Mutation(s): 2 Gene Names: sodC EC: 1.15.1.1 | 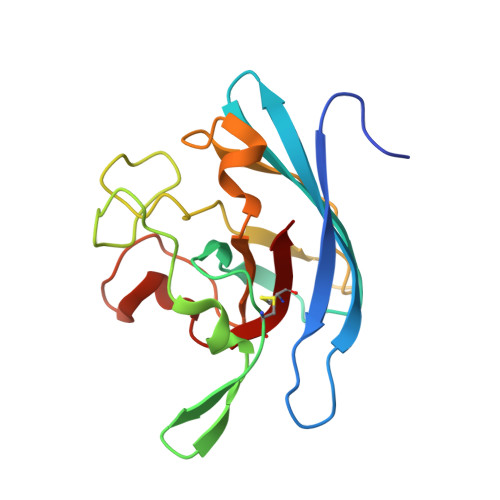 | |
UniProt | |||||
Find proteins for Q59623 (Neisseria meningitidis serogroup B (strain ATCC BAA-335 / MC58)) Explore Q59623 Go to UniProtKB: Q59623 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q59623 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | D [auth A], G [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CU Query on CU | E [auth A] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| CU1 Query on CU1 | C [auth A], F [auth B] | COPPER (I) ION Cu VMQMZMRVKUZKQL-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 45.787 | α = 90 |
| b = 66.492 | β = 101.48 |
| c = 49.752 | γ = 90 |
| Software Name | Purpose |
|---|---|
| CNS | refinement |
| MOSFLM | data reduction |
| CCP4 | data scaling |
| AMoRE | phasing |