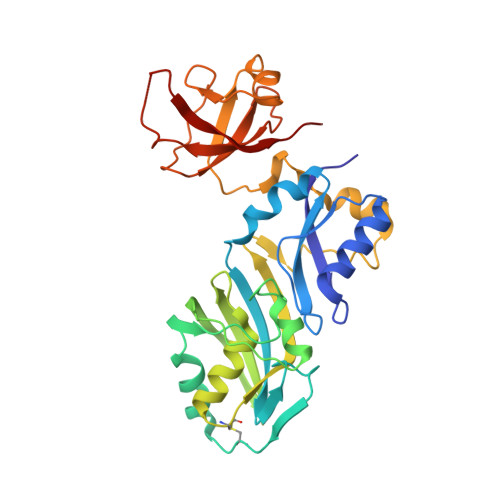
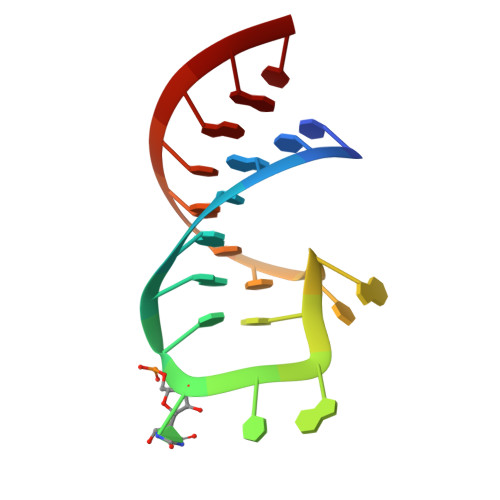
Dissecting the roles of a strictly conserved tyrosine in substrate recognition and catalysis by pseudouridine 55 synthase.
Phannachet, K., Elias, Y., Huang, R.H.(2005) Biochemistry 44: 15488-15494
- PubMed: 16300397
- DOI: https://doi.org/10.1021/bi050961w
- Primary Citation of Related Structures:
2AB4 - PubMed Abstract:
Sequence alignment of the TruA, TruB, RsuA, and RluA families of pseudouridine synthases (PsiS) identifies a strictly conserved aspartic acid, which has been shown to be the critical nucleophile for the PsiS-catalyzed formation of pseudouridine (Psi). However, superposition of the representative structures from these four families of enzymes identifies two additional amino acids, a lysine or an arginine (K/R) and a tyrosine (Y), from a K/RxY motif that are structurally conserved in the active site. We have created a series of Thermotoga maritima and Escherichia coli pseudouridine 55 synthase (Psi55S) mutants in which the conserved Y is mutated to other amino acids. A new crystal structure of the T. maritima Psi55S Y67F mutant in complex with a 5FU-RNA at 2.4 A resolution revealed formation of 5-fluoro-6-hydroxypseudouridine (5FhPsi), the same product previously seen in wild-type Psi55S-5FU-RNA complex structures. HPLC analysis confirmed efficient formation of 5FhPsi by both Psi55S Y67F and Y67L mutants but to a much lesser extent by the Y67A mutant when 5FU-RNA substrate was used. However, both HPLC analysis and a tritium release assay indicated that these mutants had no detectable enzymatic activity when the natural RNA substrate was used. The combined structural and mutational studies lead us to propose that the side chain of the conserved tyrosine in these four families of PsiS plays a dual role within the active site, maintaining the structural integrity of the active site through its hydrophobic phenyl ring and acting as a general base through its OH group for the proton abstraction required in the last step of PsiS-catalyzed formation of Psi.
- Department of Biochemistry, School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, Illinois 61801, USA.
Organizational Affiliation: