Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action
Baram, D., Pyetan, E., Sittner, A., Auerbach-Nevo, T., Bashan, A., Yonath, A.(2005) Proc Natl Acad Sci U S A 102: 12017-12022
- PubMed: 16091460
- DOI: https://doi.org/10.1073/pnas.0505581102
- Primary Citation of Related Structures:
2AAR - PubMed Abstract:
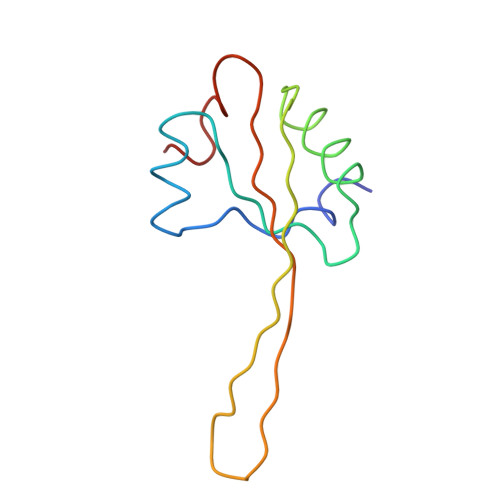
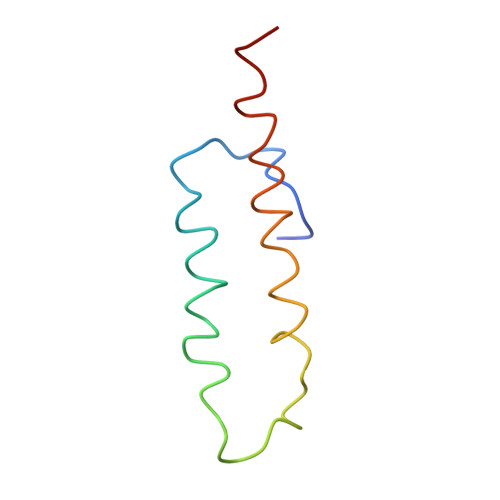
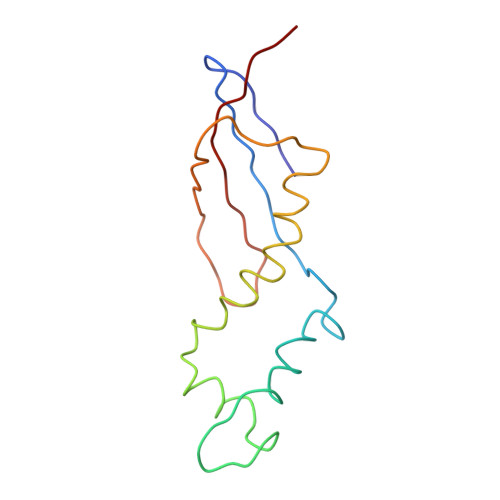
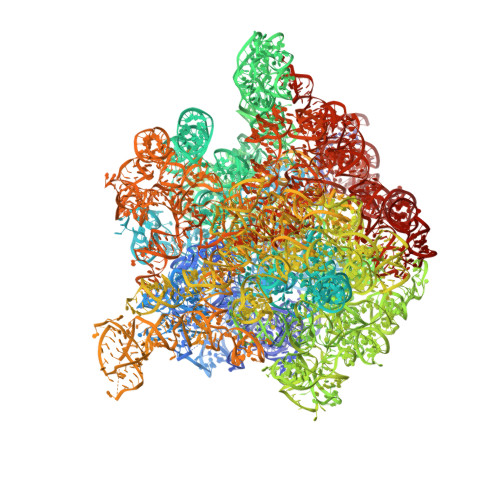
Trigger factor (TF), the first chaperone in eubacteria to encounter the emerging nascent chain, binds to the large ribosomal subunit in the vicinity of the protein exit tunnel opening and forms a sheltered folding space. Here, we present the 3.5-A crystal structure of the physiological complex of the large ribosomal subunit from the eubacterium Deinococcus radiodurans with the N-terminal domain of TF (TFa) from the same organism. For anchoring, TFa exploits a small ribosomal surface area in the vicinity of proteins L23 and L29, by using its "signature motif" as well as additional structural elements. The molecular details of TFa interactions reveal that L23 is essential for the association of TF with the ribosome and may serve as a channel of communication with the nascent chain progressing in the tunnel. L29 appears to induce a conformational change in TFa, which results in the exposure of TFa hydrophobic patches to the opening of the ribosomal exit tunnel, thus increasing its affinity for hydrophobic segments of the emerging nascent polypeptide. This observation implies that, in addition to creating a protected folding space for the emerging nascent chain, TF association with the ribosome prevents aggregation by providing a competing hydrophobic environment and may be critical for attaining the functional conformation necessary for chaperone activity.
- Department of Structural Biology, The Weizmann Institute of Science, Rehovot 76100, Israel.
Organizational Affiliation: