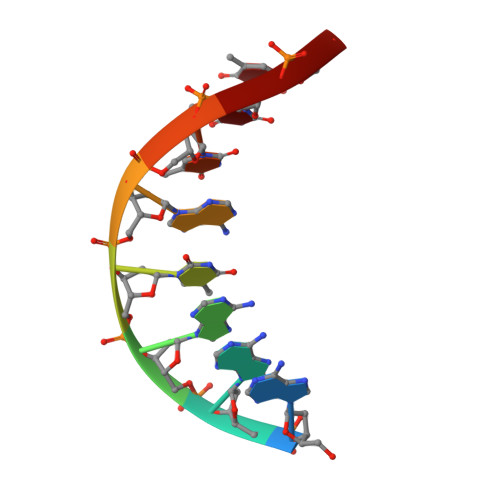
Structure of a DNA duplex with all-AT base pairs.
Valls, N., Richter, M., Subirana, J.A.(2005) Acta Crystallogr D Biol Crystallogr 61: 1587-1593
- PubMed: 16301792
- DOI: https://doi.org/10.1107/S0907444905029781
- Primary Citation of Related Structures:
2A2T - PubMed Abstract:
The structure of the oligonucleotide duplex d(AAATATTT)(2) has been obtained in two crystal forms. In both cases the duplexes show Watson-Crick base pairs and are organized as a helical arrangement of stacked oligonucleotides. The helices contain either 11 duplexes in seven turns (form I) or 14 duplexes in nine turns (form II). As a result, the crystals have rather large unit cells. Such an organization of DNA duplexes has not been previously described in oligonucleotide crystal structures. The columns of stacked duplexes are organized in a pseudohexagonal arrangement. They do not show any direct lateral interactions; instead, these are indirectly mediated by shared and disordered hydrated ions. Such interactions are similar to those found in DNA fibres. The structural parameters of the individual base steps coincide with those found in other structures which contain CG terminal base pairs.
- Departament d'Enginyeria Química, ETSEIB, Universitat Politècnica de Catalunya, Barclelona, Spain.
Organizational Affiliation: