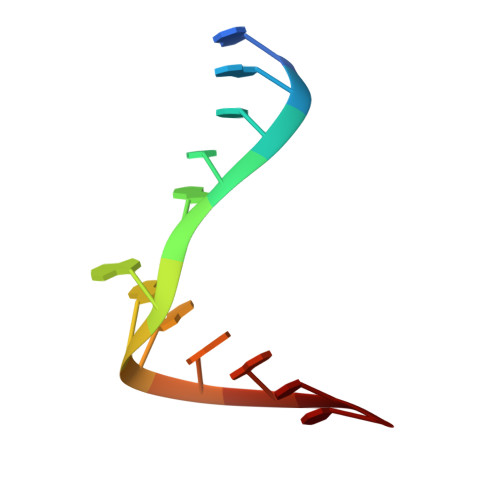
A curved RNA helix incorporating an internal loop with G.A and A.A non-Watson-Crick base pairing.
Baeyens, K.J., De Bondt, H.L., Pardi, A., Holbrook, S.R.(1996) Proc Natl Acad Sci U S A 93: 12851-12855
- PubMed: 8917508
- DOI: https://doi.org/10.1073/pnas.93.23.12851
- Primary Citation of Related Structures:
283D - PubMed Abstract:
The crystal structure of the RNA dodecamer 5'-GGCC(GAAA)GGCC-3' has been determined from x-ray diffraction data to 2.3-A resolution. In the crystal, these oligomers form double helices around twofold symmetry axes. Four consecutive non-Watson-Crick base pairs make up an internal loop in the middle of the duplex, including sheared G.A pairs and novel asymmetric A.A pairs. This internal loop sequence produces a significant curvature and narrowing of the double helix. The helix is curved by 34 degrees from end to end and the diameter is narrowed by 24% in the internal loop. A Mn2+ ion is bound directly to the N7 of the first guanine in the Watson-Crick region following the internal loop and the phosphate of the preceding residue. This Mn2+ location corresponds to a metal binding site observed in the hammerhead catalytic RNA.
- Structural Biology Division, Lawrence Berkeley National Laboratory, University of California, Berkeley 94720, USA.
Organizational Affiliation: