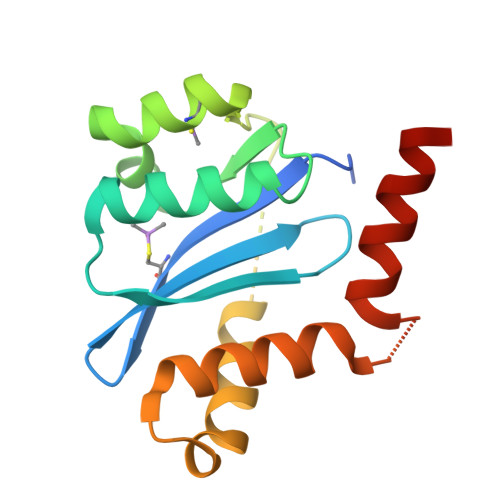
Discovery and optimization of novel and potent allosteric HIV-1 integrase inhibitors with a spiro[indene] moiety
Adachi, K., Manabe, T., Yamasaki, T., Suma, A., Oghoshi, Y., Takahashi, A., Orita, T., Nomura, A., Adachi, T., Ohata, Y., Akiyama, Y., Miyazaki, S.To be published.