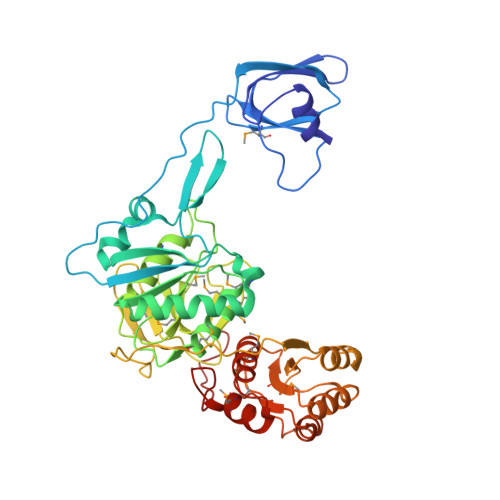
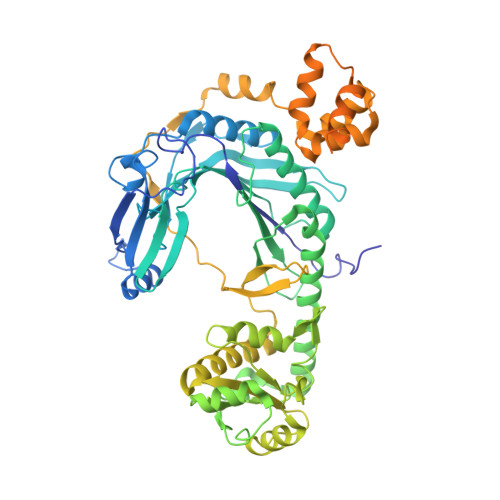
Structural Basis for tRNA-Dependent Amidotransferase Function
Schmitt, E., Panvert, M., Blanquet, S., Mechulam, Y.(2005) Structure 13: 1421-1433
- PubMed: 16216574
- DOI: https://doi.org/10.1016/j.str.2005.06.016
- Primary Citation of Related Structures:
1ZQ1 - PubMed Abstract:
Besides direct charging of tRNAs by aminoacyl-tRNA synthetases, indirect routes also ensure attachment of some amino acids onto tRNA. Such routes may explain how new amino acids entered into protein synthesis. In archaea and in most bacteria, tRNA(Gln) is first misaminoacylated by glutamyl-tRNA synthetase. Glu-tRNA(Gln) is then matured into Gln-tRNA(Gln) by a tRNA-dependent amidotransferase. We report the structure of a tRNA-dependent amidotransferase-that of GatDE from Pyrococcus abyssi. The 3.0 A resolution crystal structure shows a tetramer with two GatD molecules as the core and two GatE molecules at the periphery. The fold of GatE cannot be related to that of any tRNA binding enzyme. The ammonium donor site on GatD and the tRNA site on GatE are markedly distant. Comparison of GatD and L-asparaginase structures shows how the motion of a beta hairpin region containing a crucial catalytic threonine may control the overall reaction cycle of GatDE.
- Laboratoire de Biochimie, Unité Mixte de Recherche 7654, CNRS-Ecole Polytechnique, F-91128 Palaiseau cedex, France. emma@botrytis.polytechnique.fr
Organizational Affiliation: