A General Model for Preferential Hetero-oligomerization of LIN-2/7 Domains: Mechanism Underlying Directed Assembly of Supramolecular Signaling Complexes
Petrosky, K.Y., Ou, H.D., Lohr, F., Dotsch, V., Lim, W.A.(2005) J Biological Chem 280: 38528-38536
- PubMed: 16147993
- DOI: https://doi.org/10.1074/jbc.M506536200
- Primary Citation of Related Structures:
1ZL8 - PubMed Abstract:
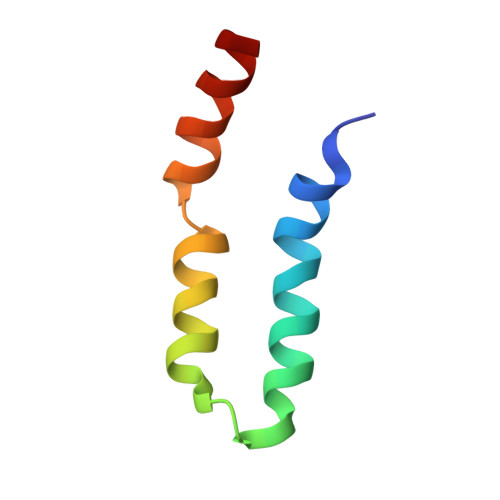
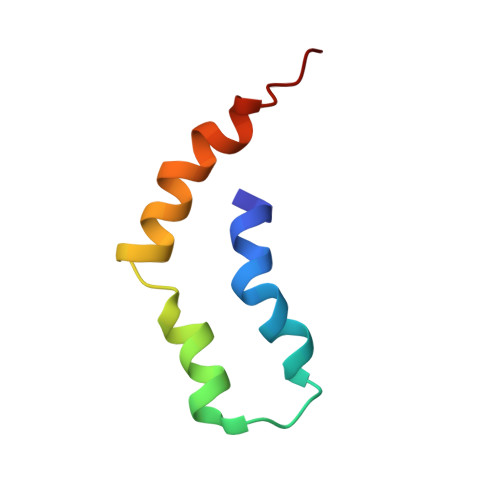
LIN-2/7 (L27) domains are protein interaction modules that preferentially hetero-oligomerize, a property critical for their function in directing specific assembly of supramolecular signaling complexes at synapses and other polarized cell-cell junctions. We have solved the solution structure of the heterodimer composed of the L27 domains from LIN-2 and LIN-7. Comparison of this structure with other L27 domain structures has allowed us to formulate a general model for why most L27 domains form an obligate heterodimer complex. L27 domains can be divided in two types (A and B), with each heterodimer comprising an A/B pair. We have identified two keystone positions that play a central role in discrimination. The residues at these positions are energetically acceptable in the context of an A/B heterodimer, but would lead to packing defects or electrostatic repulsion in the context of A/A and B/B homodimers. As predicted by the model, mutations of keystone residues stabilize normally strongly disfavored homodimers. Thus, L27 domains are specifically optimized to avoid homodimeric interactions.
- Biophysics Graduate Program, University of California, San Francisco, California 94143, USA.
Organizational Affiliation: