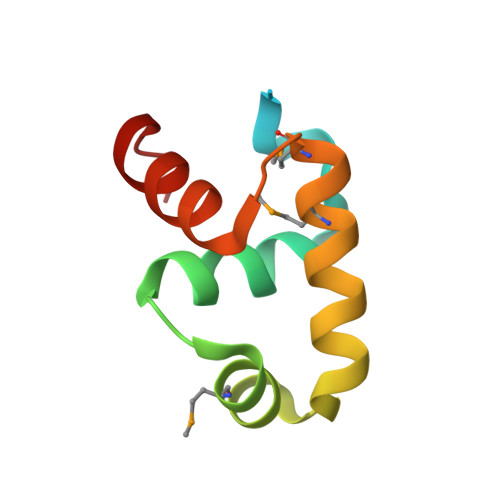
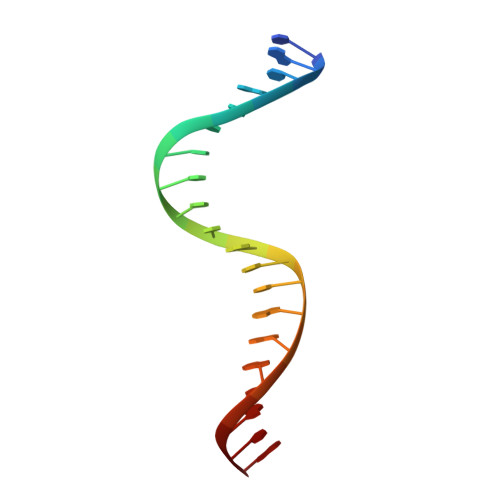
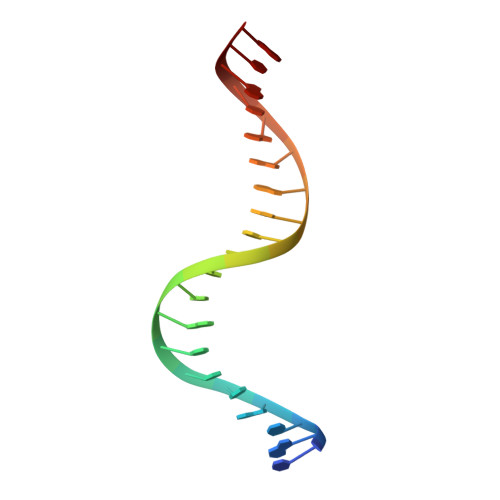
Primary and Secondary Modes of DNA Recognition by the NarL Two-Component Response Regulator.
Maris, A.E., Kaczor-Grzeskowiak, M., Ma, Z., Kopka, M.L., Gunsalus, R.P., Dickerson, R.E.(2005) Biochemistry 44: 14538-14552
- PubMed: 16262254
- DOI: https://doi.org/10.1021/bi050734u
- Primary Citation of Related Structures:
1ZG1, 1ZG5 - PubMed Abstract:
NarL is a model response regulator for bacterial two-component signal transduction. The NarL C-terminal domain DNA binding domain alone (NarL(C)) contains all essential DNA binding determinants of the full-length NarL transcription factor. In the full-length NarL protein, the N-terminal regulatory domain must be phosphorylated to release the DNA binding determinants; however, the first NarL(C)-DNA cocrystal structure showed that dimerization of NarL(C) on DNA occurs in a manner independent of the regulatory domain [Maris, A. E., et al. (2002) Nat. Struct. Biol. 9, 771-778]. Dimerization via the NarL(C) C-terminal helix conferred high-affinity recognition of the tail-to-tail promoter site arrangement. Here, two new cocrystal structures are presented of NarL(C) complexed with additional 20mer oligonucleotides representative of other high-affinity tail-to-tail NarL binding sites found in upstream promoter regions. DNA structural recognition properties are described, such as backbone flexibility and groove width, that facilitate NarL(C) dimerization and high-affinity recognition. Lys 188 on the recognition helix accommodates DNA sequence variation between the three different cocomplexes by providing flexible specificity, recognizing the DNA major groove floor directly and/or via bridging waters. The highly conserved Val 189, which enforced significant DNA base distortion in the first cocrystal structure, enforces similar distortions in the two new cocrystal structures. Recognition also is conserved for Lys 192, which hydrogen bonds to guanines at regions of high DNA helical writhe. DNA affinity measurements for model NarL binding sites, including those that did not cocrystallize, suggest a framework for explaining the diversity of heptamer site arrangement and orientation.
- Department of Chemistry and Biochemistry, University of California, Los Angeles, California 90095, USA. aemaris@lbl.gov
Organizational Affiliation: