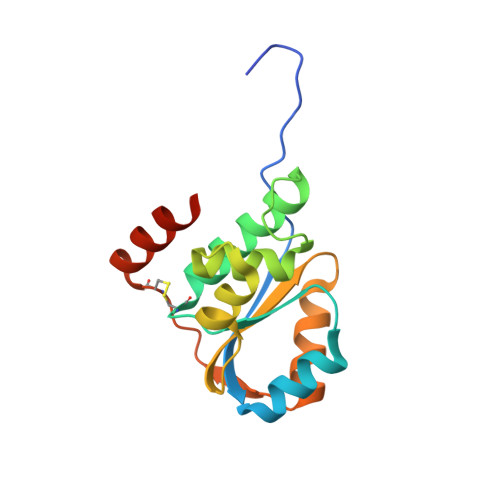
Crystal structure of Pfu-542154 conserved hypothetical protein
Habel, J.E., Liu, Z.J., Horanyi, P.S., Florence, Q.J.T., Tempel, W., Zhou, W., Chen, L., Lee, D., Nguyen, J., Chang, S.H., Bereton, P., Izumi, M., Jenny Jr., F.E., Poole II, F.L., Shah, C., Sugar, F.J., Adams, M.W.W., Rose, J.P., Wang, B.C.To be published.