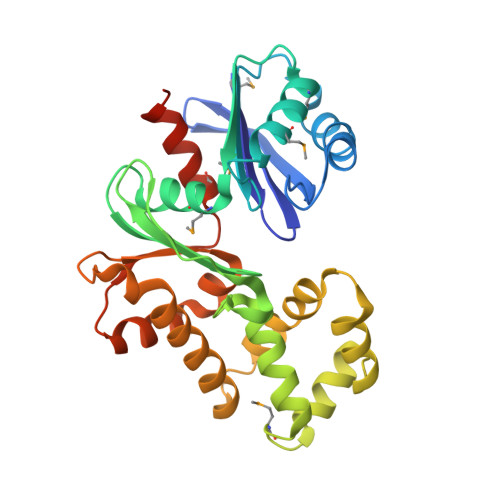
Crystal Structure of the Putative N-acetylglucosamine Kinase (PG1100) from Porphyromonas gingivalis, Northeast Structural Genomics Target PgR18
Forouhar, F., Abashidze, M., Kuzin, A., Vorobiev, S.M., Conover, K., Acton, T.B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.