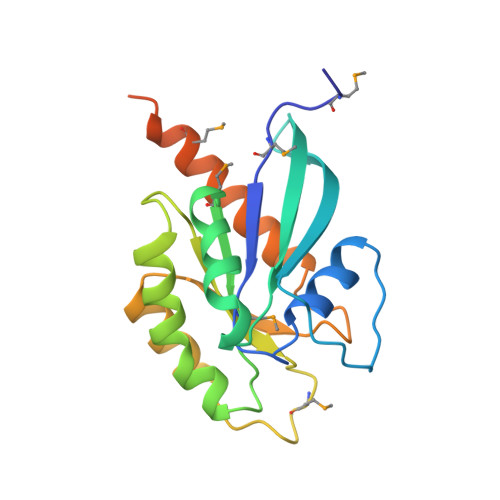
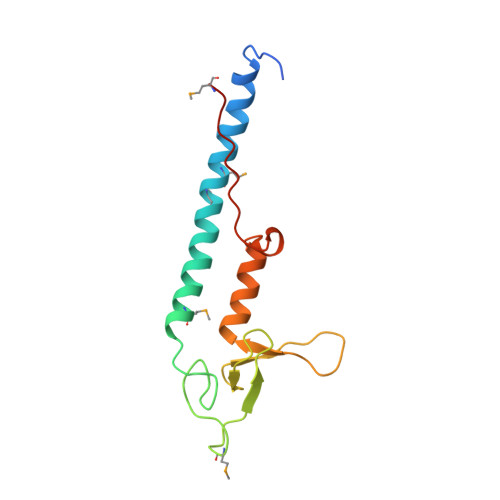
Structural basis of Rab effector specificity: crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A.
Ostermeier, C., Brunger, A.T.(1999) Cell 96: 363-374
- PubMed: 10025402
- DOI: https://doi.org/10.1016/s0092-8674(00)80549-8
- Primary Citation of Related Structures:
1ZBD - PubMed Abstract:
The small G protein Rab3A plays an important role in the regulation of neurotransmitter release. The crystal structure of activated Rab3A/GTP/Mg2+ bound to the effector domain of rabphilin-3A was solved to 2.6 A resolution. Rabphilin-3A contacts Rab3A in two distinct areas. The first interface involves the Rab3A switch I and switch II regions, which are sensitive to the nucleotide-binding state of Rab3A. The second interface consists of a deep pocket in Rab3A that interacts with a SGAWFF structural element of rabphilin-3A. Sequence and structure analysis, and biochemical data suggest that this pocket, or Rab complementarity-determining region (RabCDR), establishes a specific interaction between each Rab protein and its effectors. RabCDRs could be major determinants of effector specificity during vesicle trafficking and fusion.
- The Howard Hughes Medical Institute and Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut 06520, USA.
Organizational Affiliation: