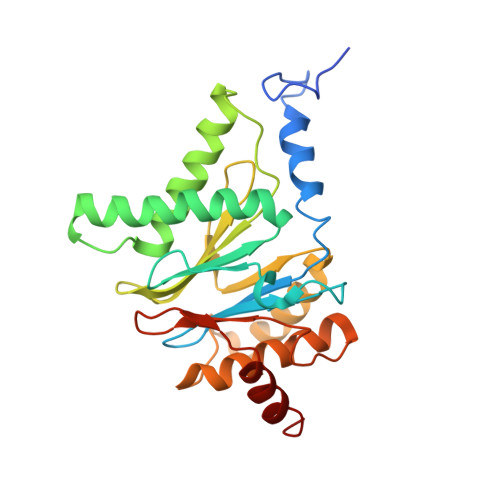
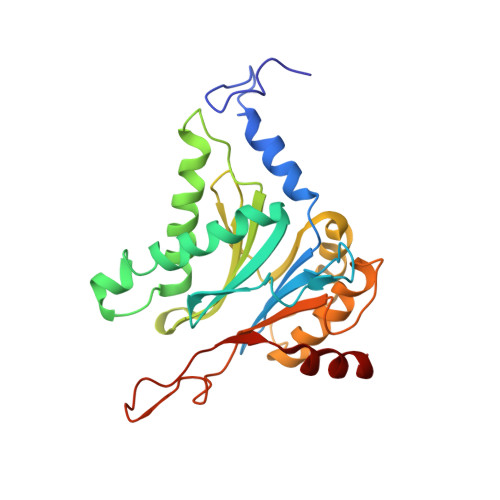
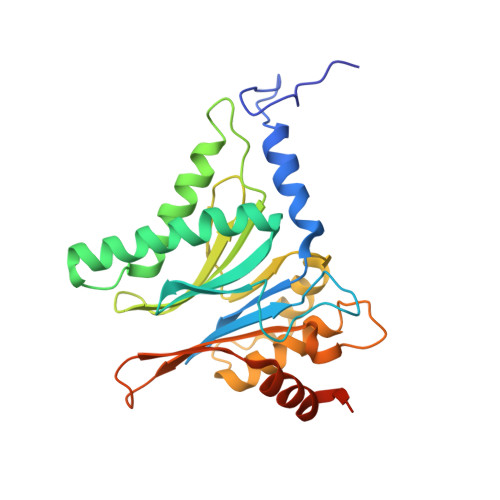
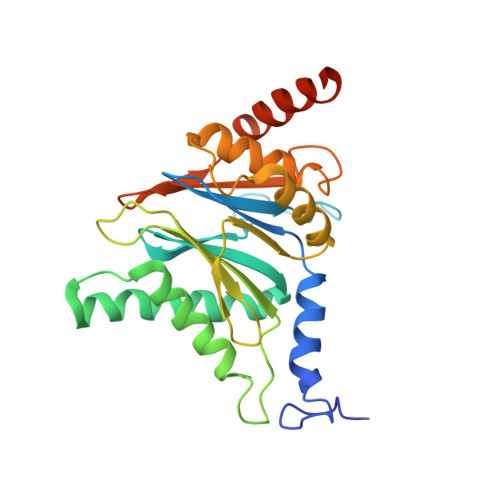
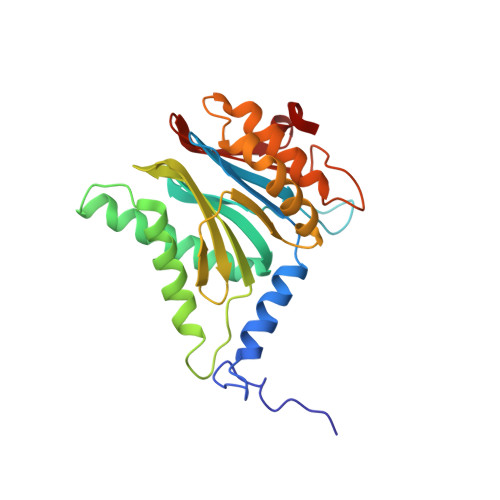
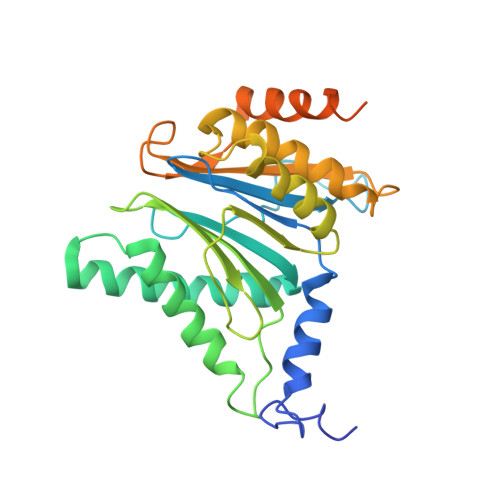
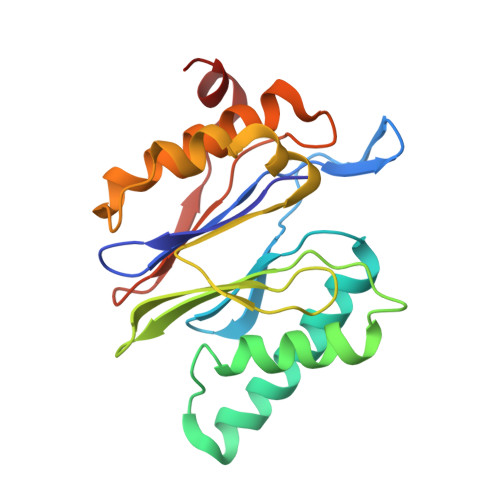
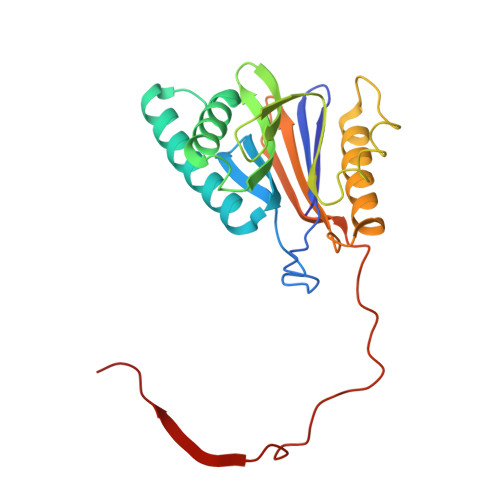
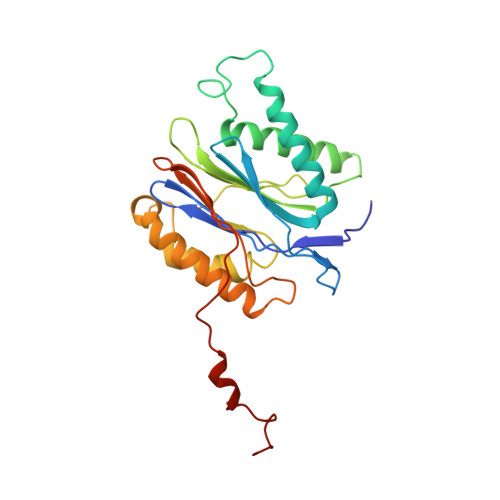
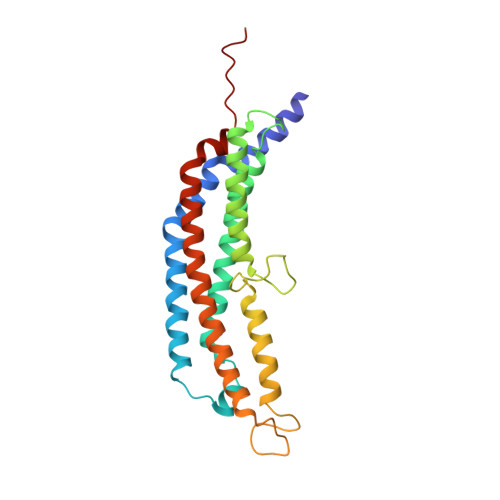
The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions.
Forster, A., Masters, E.I., Whitby, F.G., Robinson, H., Hill, C.P.(2005) Mol Cell 18: 589-599
- PubMed: 15916965
- DOI: https://doi.org/10.1016/j.molcel.2005.04.016
- Primary Citation of Related Structures:
1YA7, 1YAR, 1YAU, 1Z7Q - PubMed Abstract:
Proteasomes are cylindrical structures that function in multiple cellular processes by degrading a wide variety of cytosolic and nuclear proteins. Substrate access and product release from the enclosed catalytic chamber occurs through axial pores that are opened by activator complexes. Here, we report high-resolution structures of wild-type and mutant archaeal proteasomes bound to the activator PA26. These structures support the proposal that an ordered open conformation is required for proteolysis and that its formation can be triggered by outward displacement of surrounding residues. The structures and associated biochemical assays reveal the mechanism of binding, which involves an interaction between the PA26 C terminus and a conserved lysine. Surprisingly, biochemical observations implicate an equivalent interaction for the unrelated ATP-dependent activators PAN and PA700.
- Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, Utah 84132, USA.
Organizational Affiliation: