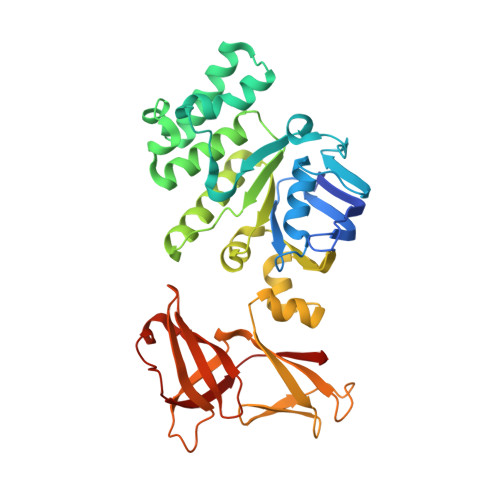
Structure of the ATPase subunit CysA of the putative sulfate ATP-binding cassette (ABC) transporter from Alicyclobacillus acidocaldarius
Scheffel, F., Demmer, U., Warkentin, E., Schneider, E., Ermler, U.(2005) FEBS Lett 579: 2953-2958
- PubMed: 15893314
- DOI: https://doi.org/10.1016/j.febslet.2005.04.017
- Primary Citation of Related Structures:
1Z47 - PubMed Abstract:
CysA, the ATPase subunit of a putative sulfate ATP-binding cassette transport system of the gram-positive thermoacidophilic bacterium Alicyclobacillus acidocaldarius, was structurally characterized at a resolution of 2.0 Angstroms in the absence of nucleotides. In line with previous findings on ABC-ATPases the structures of the two monomers (called CysA-1 and CysA-2) in the asymmetric unit differ substantially in the arrangement of their individual (sub)domains. CysA-2 was found as a physiological dimer composed of two crystallographically related monomers that are arranged in an open state. Interestingly, while the regulatory domain of CysA-2 packs against its opposing domain that of CysA-1 undergoes a conformational change and, in the dimer, would interfere with the opposing monomer thereby preventing solute translocation. Whether this conformational state is used for regulatory purposes will be discussed.
- Humboldt-Universität zu Berlin, Mathematisch-Naturwissenschaftliche Fakultät I, Institut für Biologie, Germany.
Organizational Affiliation: