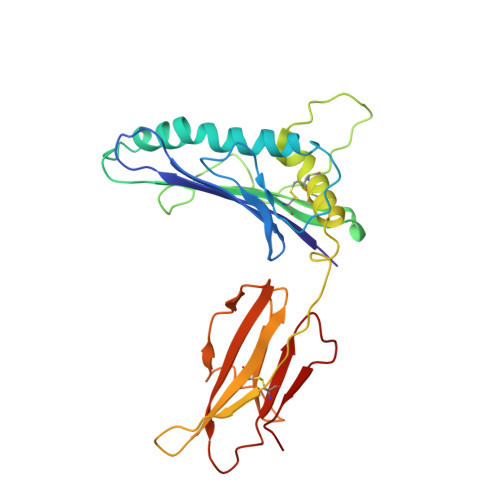
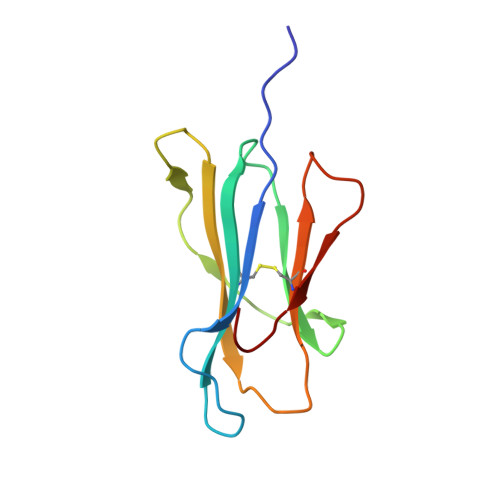
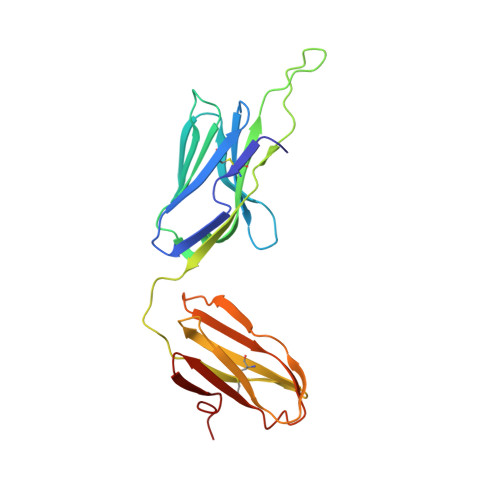
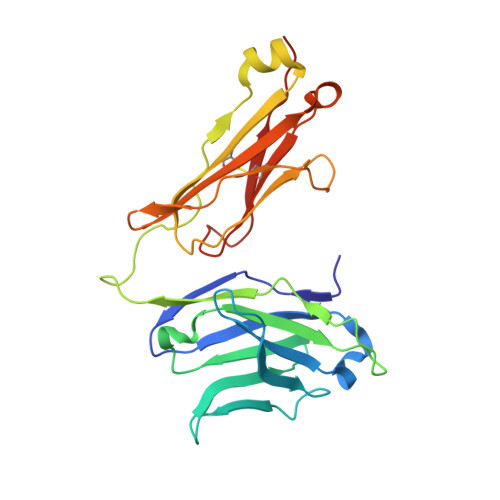
Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22.
Adams, E.J., Chien, Y.H., Garcia, K.C.(2005) Science 308: 227-231
- PubMed: 15821084
- DOI: https://doi.org/10.1126/science.1106885
- Primary Citation of Related Structures:
1YPZ - PubMed Abstract:
Gammadelta T cell receptors (TCRs), alphabeta TCRs, and antibodies are the three lineages of somatically recombined antigen receptors. The structural basis for ligand recognition is well defined for alphabeta TCR and antibodies but is lacking for gammadelta TCRs. We present the 3.4 A structure of the murine gammadelta TCR G8 bound to its major histocompatibility complex (MHC) class Ib ligand, T22. G8 predominantly uses germline-encoded residues of its delta chain complementarity-determining region 3 (CDR3) loop to bind T22 in an orientation substantially different from that seen in alphabeta TCR/peptide-MHC. That junctionally encoded G8 residues play an ancillary role in binding suggests a fusion of innate and adaptive recognition strategies.
- Department of Microbiology and Immunology, Stanford University School of Medicine, Fairchild D319, 299 Campus Drive, Stanford, CA 94035-5124, USA.
Organizational Affiliation: