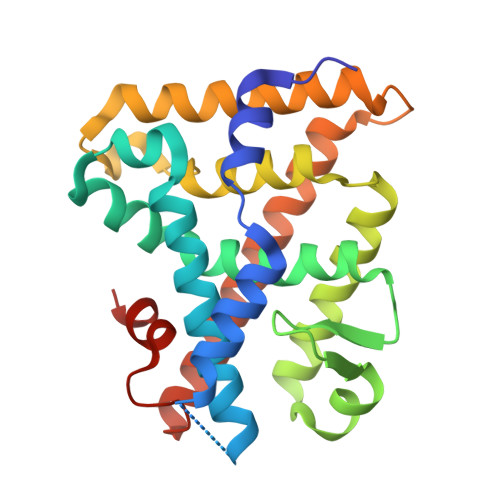
Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1
Krylova, I.N., Sablin, E.P., Xu, R.X., Waitt, G.M., MacKay, J.A., Juzumiene, D., Bynum, J.M., Madauss, K.P., Montana, V., Lebedeva, L., Suzawa, M., Williams, J.D., Williams, S.P., Guy, R.K., Thornton, J.W., Fletterick, R.J., Willson, T.M., Ingraham, H.A.(2005) Cell 120: 343-355
- PubMed: 15707893
- DOI: https://doi.org/10.1016/j.cell.2005.01.024
- Primary Citation of Related Structures:
1YMT, 1YOK, 1YOW - PubMed Abstract:
Vertebrate members of the nuclear receptor NR5A subfamily, which includes steroidogenic factor 1 (SF-1) and liver receptor homolog 1 (LRH-1), regulate crucial aspects of development, endocrine homeostasis, and metabolism. Mouse LRH-1 is believed to be a ligand-independent transcription factor with a large and empty hydrophobic pocket. Here we present structural and biochemical data for three other NR5A members-mouse and human SF-1 and human LRH-1-which reveal that these receptors bind phosphatidyl inositol second messengers and that ligand binding is required for maximal activity. Evolutionary analysis of structure-function relationships across the SF-1/LRH-1 subfamily indicates that ligand binding is the ancestral state of NR5A receptors and was uniquely diminished or altered in the rodent LRH-1 lineage. We propose that phospholipids regulate gene expression by directly binding to NR5A nuclear receptors.
- Department of Physiology, University of California, San Francisco, California 94143, USA.
Organizational Affiliation: