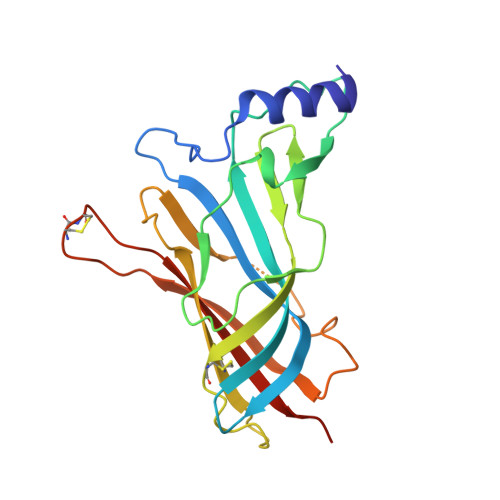
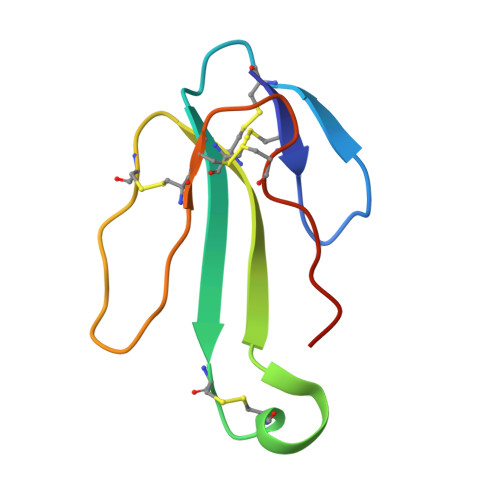
Crystal structure of a Cbtx-AChBP complex reveals essential interactions between snake alpha-neurotoxins and nicotinic receptors
Bourne, Y., Talley, T.T., Hansen, S.B., Taylor, P., Marchot, P.(2005) EMBO J 24: 1512-1522
- PubMed: 15791209
- DOI: https://doi.org/10.1038/sj.emboj.7600620
- Primary Citation of Related Structures:
1YI5 - PubMed Abstract:
The crystal structure of the snake long alpha-neurotoxin, alpha-cobratoxin, bound to the pentameric acetylcholine-binding protein (AChBP) from Lymnaea stagnalis, was solved from good quality density maps despite a 4.2 A overall resolution. The structure unambiguously reveals the positions and orientations of all five three-fingered toxin molecules inserted at the AChBP subunit interfaces and the conformational changes associated with toxin binding. AChBP loops C and F that border the ligand-binding pocket move markedly from their original positions to wrap around the tips of the toxin first and second fingers and part of its C-terminus, while rearrangements also occur in the toxin fingers. At the interface of the complex, major interactions involve aromatic and aliphatic side chains within the AChBP binding pocket and, at the buried tip of the toxin second finger, conserved Phe and Arg residues that partially mimic a bound agonist molecule. Hence this structure, in revealing a distinctive and unpredicted conformation of the toxin-bound AChBP molecule, provides a lead template resembling a resting state conformation of the nicotinic receptor and for understanding selectivity of curaremimetic alpha-neurotoxins for the various receptor species.
- Architecture et Fonction des Macromolécules Biologiques, CNRS UMR-6098, Marseille, France. yves@afmb.cnrs-mrs.fr
Organizational Affiliation: