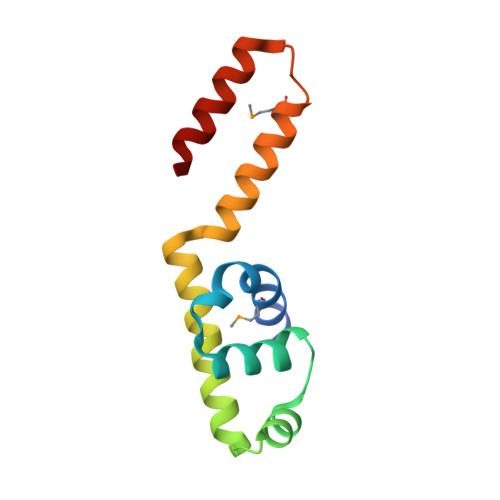
1.7 Angstrom Crystal Structure of Conserved Hypothetical UPF0122 Protein SAV1236 From Staphylococcus aureus
Walker, J.R., Xu, X., Virag, C., McDonald, M.-L., Houston, S., Buzadzija, K., Vedadi, M., Dharamsi, A., Fiebig, K.M., Savchenko, A.To be published.