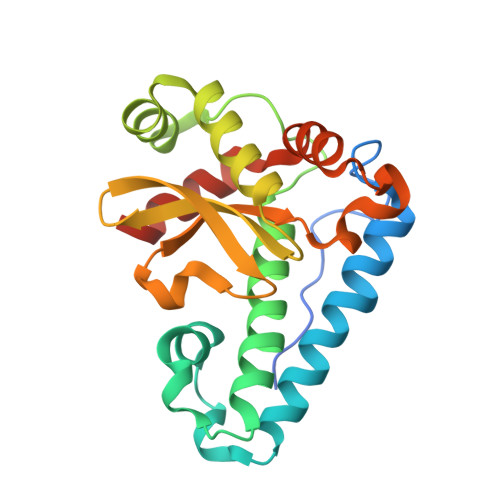
Structures of two superoxide dismutases from Bacillus anthracis reveal a novel active centre.
Boucher, I.W., Kalliomaa, A.K., Levdikov, V.M., Blagova, E.V., Fogg, M.J., Brannigan, J.A., Wilson, K.S., Wilkinson, A.J.(2005) Acta Crystallogr Sect F Struct Biol Cryst Commun 61: 621-624
- PubMed: 16511113
- DOI: https://doi.org/10.1107/S1744309105017380
- Primary Citation of Related Structures:
1XRE, 1XUQ - PubMed Abstract:
The BA4499 and BA5696 genes of Bacillus anthracis encode proteins homologous to manganese superoxide dismutase, suggesting that this organism has an expanded repertoire of antioxidant proteins. Differences in metal specificity and quaternary structure between the dismutases of prokaryotes and higher eukaryotes may be exploited in the development of therapeutic antibacterial compounds. Here, the crystal structure of two Mn superoxide dismutases from B. anthracis solved to high resolution are reported. Comparison of their structures reveals that a highly conserved residue near the active centre is substituted in one of the proteins and that this is a characteristic feature of superoxide dismutases from the B. cereus/B. anthracis/B. thuringiensis group of organisms.
- Department of Chemistry, University of York, York YO10 5YW, England.
Organizational Affiliation: