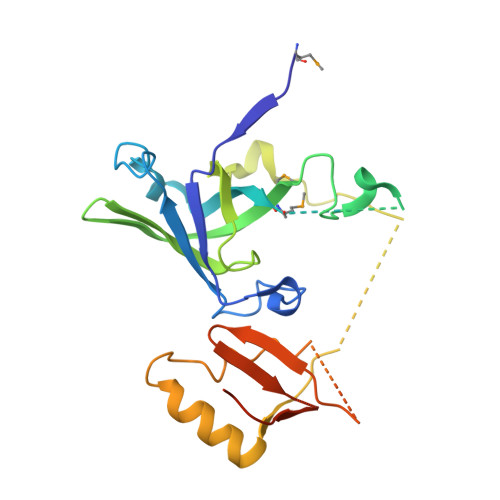
X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC) target IR47.
Benach, J., Lee, I., Forouhar, F., Kuzin, A.P., Keller, J.P., Itkin, A., Xiao, R., Acton, T., Montelione, G.T., Hunt, J.F.To be published.