The crystal structure of the bacteriophage PSA endolysin reveals a unique fold responsible for specific recognition of Listeria cell walls
Korndoerfer, I.P., Danzer, J., Schmelcher, M., Zimmer, M., Skerra, A., Loessner, M.J.(2006) J Mol Biology 364: 678-689
- PubMed: 17010991
- DOI: https://doi.org/10.1016/j.jmb.2006.08.069
- Primary Citation of Related Structures:
1XOV - PubMed Abstract:
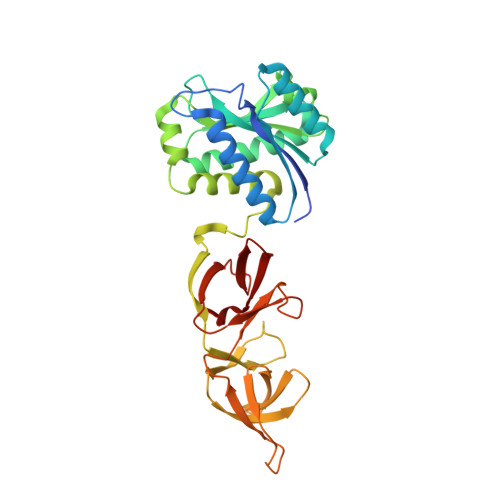
Bacteriophage murein hydrolases exhibit high specificity towards the cell walls of their host bacteria. This specificity is mostly provided by a structurally well defined cell wall-binding domain that attaches the enzyme to its solid substrate. To gain deeper insight into this mechanism we have crystallized the complete 314 amino acid endolysin from the temperate Listeria monocytogenes phage PSA. The crystal structure of PlyPSA was determined by single wavelength anomalous dispersion methods and refined to 1.8 A resolution. The two functional domains of the polypeptide, providing cell wall-binding and enzymatic activities, can be clearly distinguished and are connected via a linker segment of six amino acid residues. The core of the N-acetylmuramoyl-L-alanine amidase moiety is formed by a twisted, six-stranded beta-sheet flanked by six helices. Although the catalytic domain is unique among the known Listeria phage endolysins, its structure is highly similar to known phosphorylase/hydrolase-like alpha/beta-proteins, including an autolysin amidase from Paenibacillus polymyxa. In contrast, the C-terminal domain of PlyPSA features a novel fold, comprising two copies of a beta-barrel-like motif, which are held together by means of swapped beta-strands. The architecture of the enzyme with its two separate domains explains its unique substrate recognition properties and also provides insight into the lytic mechanisms of related Listeria phage endolysins, a class of enzymes that bear biotechnological potential.
- Technische Universität München, Lehrstuhl für Biologische Chemie, An der Saatzucht 5, D-85350 Freising, Germany. ipk@crystax.com
Organizational Affiliation: