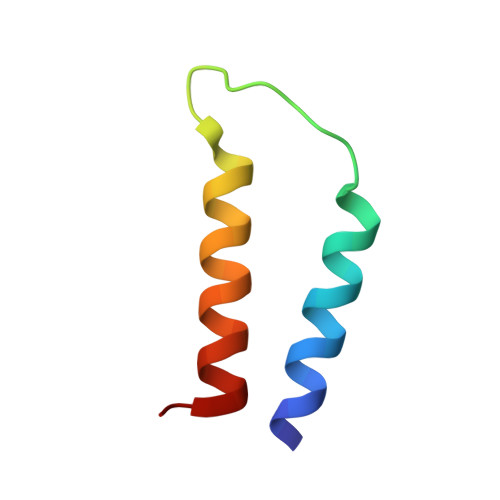
NMR Structure Determination of a Membrane Protein with Two Transmembrane Helices in Micelles: Merf of the Bacterial Mercury Detoxification System
Howell, S.C., Mesleh, M.F., Opella, S.J.(2005) Biochemistry 44: 5196
- PubMed: 15794657
- DOI: https://doi.org/10.1021/bi048095v
- Primary Citation of Related Structures:
1WAZ - PubMed Abstract:
The three-dimensional backbone structure of a membrane protein with two transmembrane helices in micelles was determined using solution NMR methods that rely on the measurement of backbone (1)H-(15)N residual dipolar couplings (RDCs) from samples of two different constructs that align differently in stressed polyacrylamide gels. Dipolar wave fitting to the (1)H-(15)N RDCs determines the helical boundaries based on periodicity and was utilized in the generation of supplemental dihedral restraints for the helical segments. The (1)H-(15)N RDCs and supplemental dihedral restraints enable the determination of the structure of the helix-loop-helix core domain of the mercury transport membrane protein MerF with a backbone RMSD of 0.58 A. Moreover, the fold of this polypeptide demonstrates that the two vicinal pairs of cysteine residues, shown to be involved in the transport of Hg(II) across the membrane, are exposed to the cytoplasm. This finding differs from earlier structural and mechanistic models that were based primarily on the somewhat atypical hydropathy plot for MerF and related transport proteins.
- Department of Chemistry and Biochemistry, University of California, San Diego, La Jolla, California 92093-0307, USA.
Organizational Affiliation: