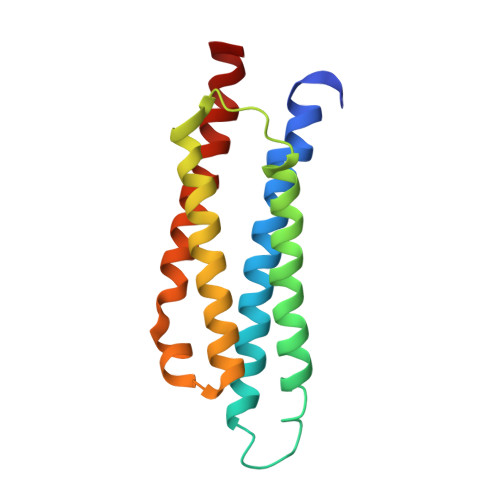
The three-dimensional structure of the ligand-binding domain of a wild-type bacterial chemotaxis receptor. Structural comparison to the cross-linked mutant forms and conformational changes upon ligand binding.
Yeh, J.I., Biemann, H.P., Pandit, J., Koshland, D.E., Kim, S.H.(1993) J Biological Chem 268: 9787-9792
- PubMed: 8486661
- Primary Citation of Related Structures:
1WAS, 1WAT - PubMed Abstract:
The three-dimensional structures of the ligand-binding domain of the wild-type Salmonella typhimurium aspartate receptor have been determined in the absence (apo) and presence of bound aspartate (complex) and compared to a cross-linked mutant containing a cysteine at position 36 which does not change signaling behavior of the intact receptor. The structures of the wild-type forms were determined in order to assess the effects of cross-linking on the structure and its influence on conformational changes upon ligand binding. As in the case of the cross-linked mutant receptor, the non-cross-linked ligand-binding domain is dimeric and is composed of 4-alpha-helical bundle monomer subunits related by a crystallographic 2-fold axis in the unbound form and by a non-crystallographic axis in the aspartate-bound form. A comparative study between the non-cross-linked and cross-linked structures has led to the following observations: 1) The long N-terminal helices of the individual subunits in the cross-linked structures are bent toward each other to accommodate the disulfide bond. 2) The rest of the subunit conformation is very similar to that of the wild-type. 3) The intersubunit angle of the cross-linked apo structure is larger by about 13 degrees when compared to the wild-type apo structure. 4) The nature and magnitude of the aspartate-induced conformational changes in the non-cross-linked wild-type structures are very similar to those of the cross-linked structures.
- Department of Chemistry, Lawrence Berkeley Laboratory, University of California, Berkeley 94720.
Organizational Affiliation: