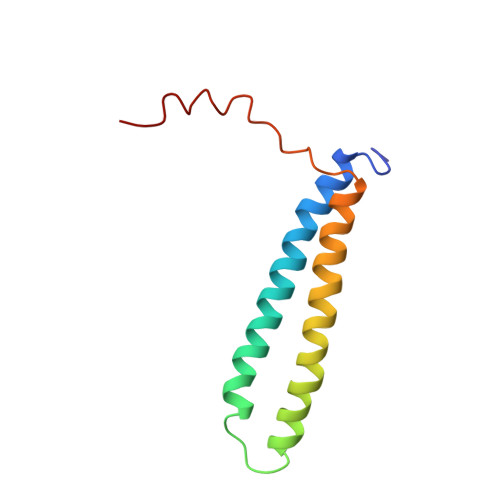
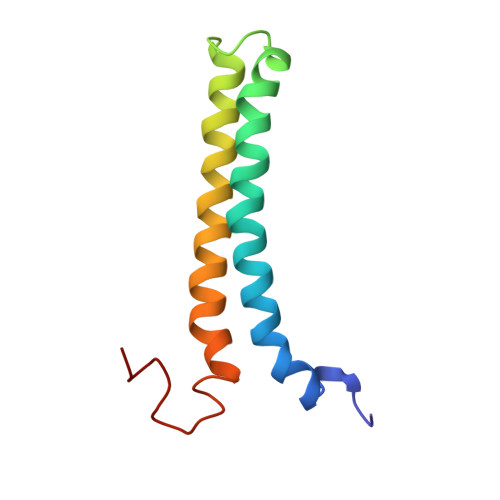
Structure and Function of the Complex Formed by the Tuberculosis Virulence Factors Cfp-10 and Esat-6
Renshaw, P.S., Lightbody, K.L., Veverka, V., Muskett, F.W., Kelly, G., Frenkiel, T.A., Gordon, S.V., Hewinson, R.G., Burke, B., Norman, J., Williamson, R.A., Carr, M.D.(2005) EMBO J 24: 2491
- PubMed: 15973432
- DOI: https://doi.org/10.1038/sj.emboj.7600732
- Primary Citation of Related Structures:
1WA8 - PubMed Abstract:
The secreted Mycobacterium tuberculosis complex proteins CFP-10 and ESAT-6 have recently been shown to play an essential role in tuberculosis pathogenesis. We have determined the solution structure of the tight, 1:1 complex formed by CFP-10 and ESAT-6, and employed fluorescence microscopy to demonstrate specific binding of the complex to the surface of macrophage and monocyte cells. A striking feature of the complex is the long flexible arm formed by the C-terminus of CFP-10, which was found to be essential for binding to the surface of cells. The surface features of the CFP-10.ESAT-6 complex, together with observed binding to specific host cells, strongly suggest a key signalling role for the complex, in which binding to cell surface receptors leads to modulation of host cell behaviour to the advantage of the pathogen.
- Department of Biochemistry, University of Leicester, Leicester, UK.
Organizational Affiliation: