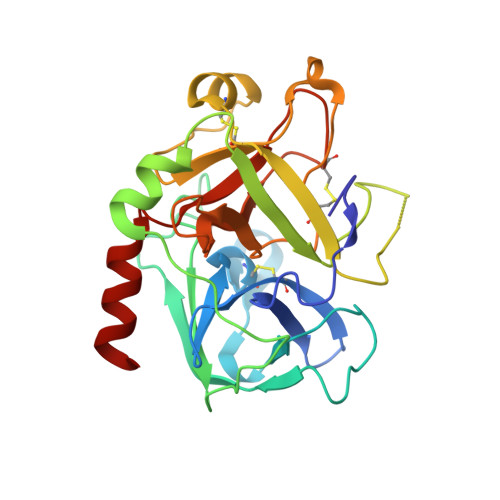
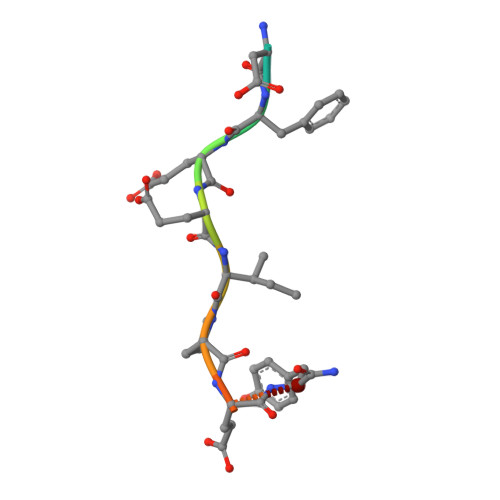
Design, Synthesis and Evaluation of Graftable Thrombin Inhibitors for the Preparation of Blood-Compatible Polymer Materials.
Salvagnini, C., Michaux, C., Remiche, J., Wouters, J., Charlier, P., Marchand-Brynaert, J.(2005) Org Biomol Chem 3: 4209
- PubMed: 16294249
- DOI: https://doi.org/10.1039/b510239a
- Primary Citation of Related Structures:
1W7G - PubMed Abstract:
Piperazinyl-amide derivatives of N-alpha-(3-trifluoromethyl-benzenesulfonyl)-L-arginine (1) were synthesized as graftable thrombin inhibitors. The possible disturbance of biological activity due to a variable spacer-arm fixed on the N-4 piperazinyl position was evaluated in vitro, against human alpha-thrombin, and in blood coagulation assay. Molecular modelling (in silico analysis) and X-ray diffraction studies of thrombin-inhibitor complexes were also performed. The fixation of bioactive molecules on poly(butylene terephthalate) (PBT) and poly(ethylene terephthalate) (PET) membranes was performed by wet chemistry treatment and evaluated by XPS analysis. Surface grafting of inhibitor 1d improved the membrane hemocompatibility by reducing blood clot formation on the modified surface.
- Unité de Chimie Organique et Médicinale, Université catholique de Louvain, Bâtiment Lavoisier, place Louis Pasteur 1, B-1348, Louvain-la-Neuve, Belgium.
Organizational Affiliation: