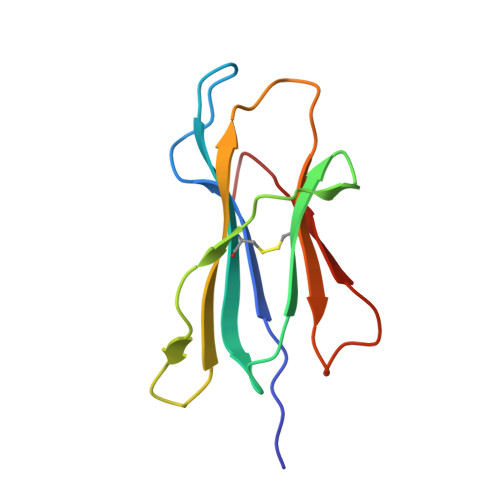
A Major Histocompatibility Complex.Peptide- Restricted Antibody and T Cell Receptor Molecules Recognize Their Target by Distinct Binding Modes: Crystal Structure of Human Leukocyte Antigen (Hla)-A1.Mage-A1 in Complex with Fab-Hyb3
Hulsmeyer, M., Chames, P., Hillig, R.C., Stanfield, R.L., Held, G., Coulie, P.G., Alings, C., Wille, G., Saenger, W., Uchanska-Ziegler, B., Hoogenboom, H.R., Ziegler, A.(2005) J Biological Chem 280: 2972
- PubMed: 15537658
- DOI: https://doi.org/10.1074/jbc.M411323200
- Primary Citation of Related Structures:
1W72 - PubMed Abstract:
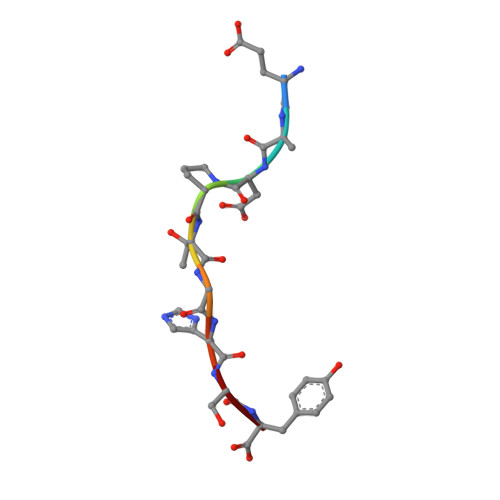
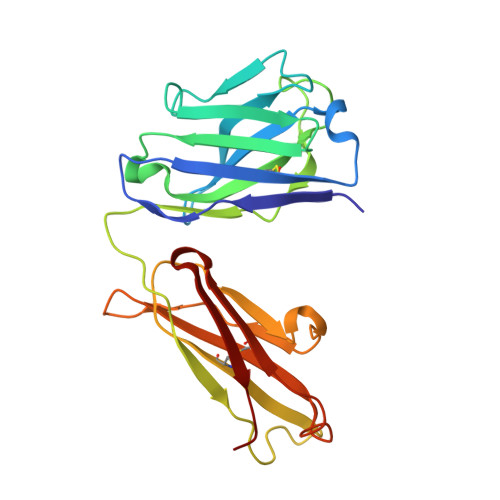
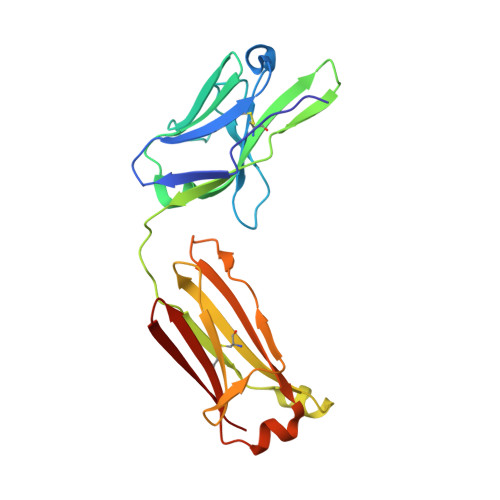
Antibodies with T cell receptor-like specificity possess a considerable diagnostic and therapeutic potential, but the structural basis of the interaction between an antibody and an histocompatibility antigen has so far not been determined. We present here the crystal structure (at 2.15 A resolution) of the recombinant, affinity-matured human antibody fragment Fab-Hyb3 bound to the tumor-associated human leukocyte antigen (HLA)/peptide complex HLA-A1.MAGE-A1. Fab-Hyb3 employs a diagonal docking mode resembling that of T cell receptors. However, other than these natural ligands, the antibody uses only four of its six complementarity-determining regions for direct interactions with the target. It recognizes the C-terminal half of the MAGE-A1 peptide, the HLA-A1 alpha1-helix, and N-terminal residues of the alpha2-helix, accompanied by a large tilting angle between the two types of molecules within the complex. Interestingly, only a single hydrogen bond between a peptide side chain and Fab-Hyb3 contributes to the interaction, but large buried surface areas with pronounced shape complementarity assure high affinity and specificity for MAGE-A1. The HLA-A1.MAGE-A1.antibody structure is discussed in comparison with those of natural ligands recognizing HLA.peptide complexes.
- Institut für Chemie/Kristallographie, Freie Universität Berlin, 14195 Berlin, Germany, Cellectis, 93235 Romainville, France.
Organizational Affiliation: