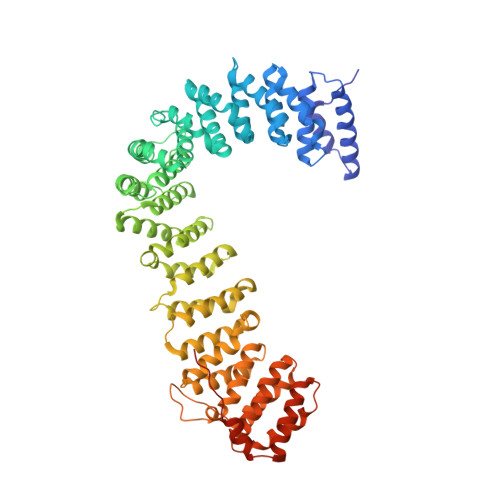
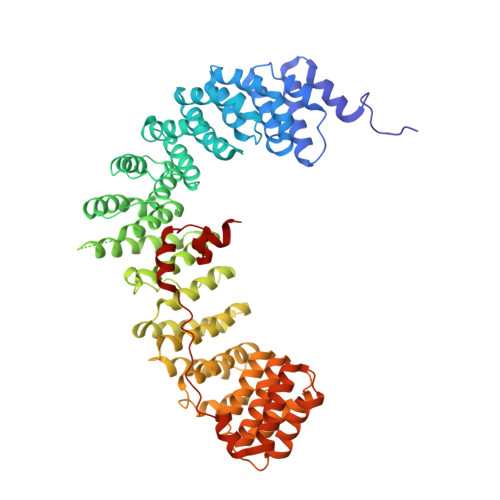
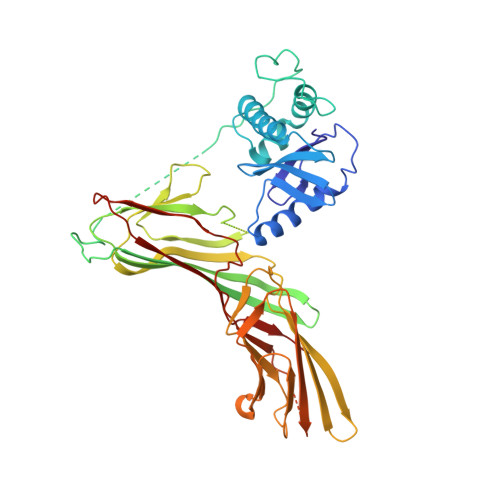
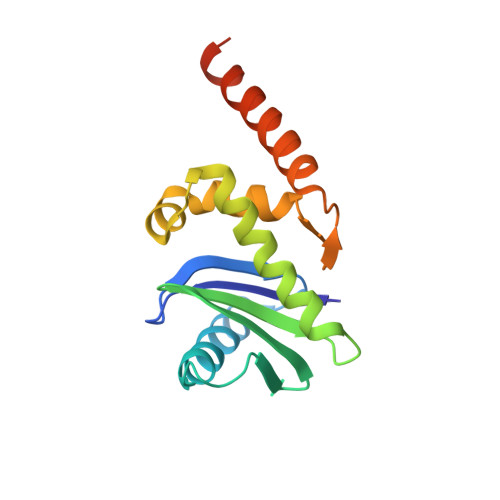
Crystal Structure of the Clathrin Adaptor Protein 1 Core
Heldwein, E., Macia, E., Wang, J., Yin, H.L., Kirchhausen, T., Harrison, S.C.(2004) Proc Natl Acad Sci U S A 101: 14108
- PubMed: 15377783
- DOI: https://doi.org/10.1073/pnas.0406102101
- Primary Citation of Related Structures:
1W63 - PubMed Abstract:
The heterotetrameric adaptor proteins (AP complexes) link the outer lattice of clathrin-coated vesicles with membrane-anchored cargo molecules. We report the crystal structure of the core of the AP-1 complex, which functions in the trans-Golgi network (TGN). Packing of complexes in the crystal generates an exceptionally long (1,135-A) unit-cell axis, but the 6-fold noncrystallographic redundancy yields an excellent map at 4-A resolution. The AP-1 core comprises N-terminal fragments of the two large chains, beta1 and gamma, and the intact medium and small chains, micro1 and sigma1. Its molecular architecture closely resembles that of the core of AP-2, the plasma-membrane-specific adaptor, for which a structure has been determined. Both structures represent an "inactive" conformation with respect to binding of cargo with a tyrosine-based sorting signal. TGN localization of AP-1 depends on the small GTPase, Arf1, and the phosphoinositide, PI-4-P. We show that directed mutations of residues at a particular corner of the gamma chain prevent recruitment to the TGN in cells and diminish PI-4-P-dependent, but not Arf1-dependent, liposome binding in vitro.
- Children's Hospital, Howard Hughes Medical Institute, Department of Cell Biology, and CBR Institute for Biomedical Research, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: