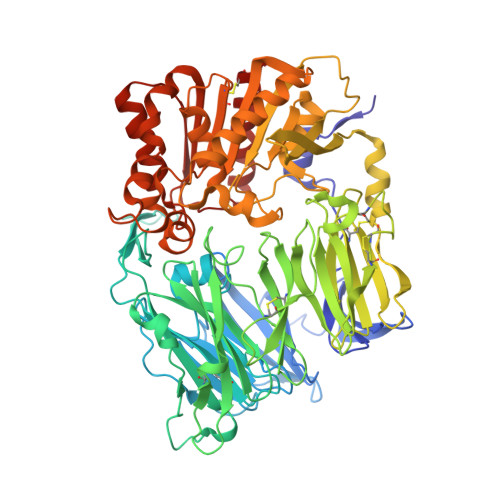
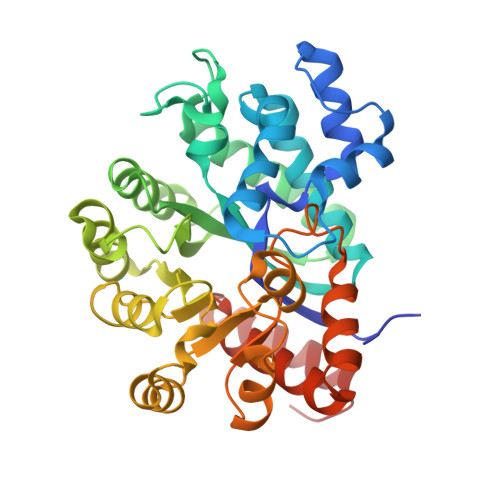
Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface.
Weihofen, W.A., Liu, J., Reutter, W., Saenger, W., Fan, H.(2004) J Biological Chem 279: 43330-43335
- PubMed: 15213224
- DOI: https://doi.org/10.1074/jbc.M405001200
- Primary Citation of Related Structures:
1W1I - PubMed Abstract:
Dipeptidyl-peptidase IV (DPPIV or CD26) is a homodimeric type II membrane glycoprotein in which the two monomers are subdivided into a beta-propeller domain and an alpha/beta-hydrolase domain. As dipeptidase, DPPIV modulates the activity of various biologically important peptides and, in addition, DPPIV acts as a receptor for adenosine deaminase (ADA), thereby mediating co-stimulatory signals in T-lymphocytes. The 3.0-A resolution crystal structure of the complex formed between human DPPIV and bovine ADA presented here shows that each beta-propeller domain of the DPPIV dimer binds one ADA. At the binding interface, two hydrophobic loops protruding from the beta-propeller domain of DPPIV interact with two hydrophilic and heavily charged alpha-helices of ADA, giving rise to the highest percentage of charged residues involved in a protein-protein contact reported thus far. Additionally, four glycosides linked to Asn229 of DPPIV bind to ADA. In the crystal structure of porcine DPPIV, the observed tetramer formation was suggested to mediate epithelial and lymphocyte cell-cell adhesion. ADA binding to DPPIV could regulate this adhesion, as it would abolish tetramerization.
- Institut für Chemie/Kristallographie, Freie Universität Berlin, Takustrasse 6, D-14195 Berlin, Germany.
Organizational Affiliation: