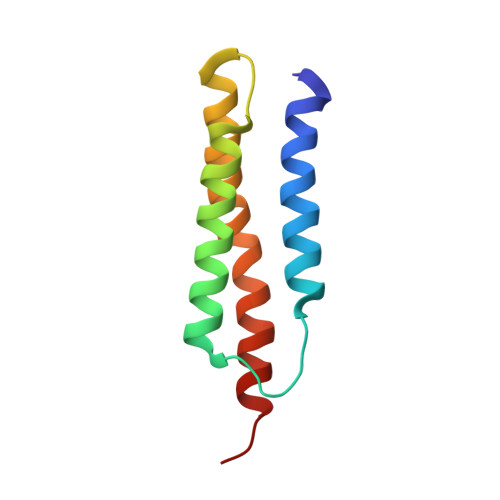
NMR structure of the alpha-hemoglobin stabilizing protein: insights into conformational heterogeneity and binding.
Santiveri, C.M., Perez-Canadillas, J.M., Vadivelu, M.K., Allen, M.D., Rutherford, T.J., Watkins, N.A., Bycroft, M.(2004) J Biological Chem 279: 34963-34970
- PubMed: 15178680
- DOI: https://doi.org/10.1074/jbc.M405016200
- Primary Citation of Related Structures:
1W09, 1W0A, 1W0B - PubMed Abstract:
The structure of alpha-hemoglobin stabilizing protein (AHSP), a molecular chaperone for free alpha-hemoglobin, has been determined using NMR spectroscopy. The protein native state shows conformational heterogeneity attributable to the isomerization of the peptide bond preceding a conserved proline residue. The two equally populated cis and trans forms both adopt an elongated antiparallel three alpha-helix bundle fold but display major differences in the loop between the first two helices and at the C terminus of helix 3. Proline to alanine single point mutation of the residue Pro-30 prevents the cis/trans isomerization. The structure of the P30A mutant is similar to the structure of the trans form of AHSP in the loop 1 region. Both the wild-type AHSP and the P30A mutant bind to alpha-hemoglobin, and the wild-type conformational heterogeneity is quenched upon complex formation, suggesting that just one conformation is the active form. Changes in chemical shift observed upon complex formation identify a binding interface comprising the C terminus of helix 1, the loop 1, and the N terminus of helix 2, with the exposed residues Phe-47 and Tyr-51 being attractive targets for molecular recognition. The characteristics of this interface suggest that AHSP binds at the intradimer alpha1beta1 interface in tetrameric HbA.
- Medical Research Council Centre for Protein Engineering, Hills Road, Cambridge CB2 2QH, United Kingdom.
Organizational Affiliation: