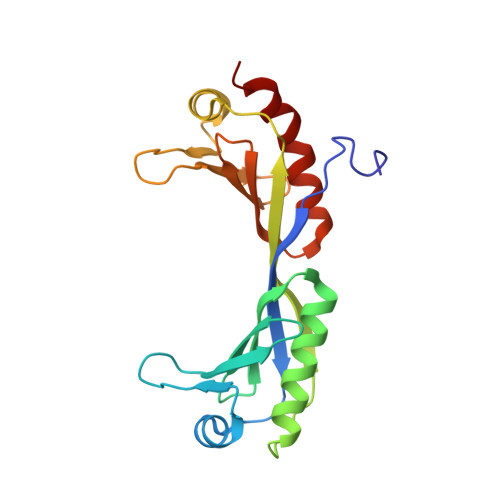
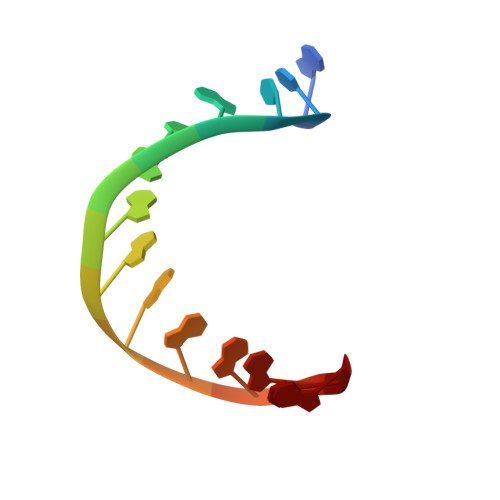
Co-Crystal Structure of TBP Recognizing the Minor Groove of a TATA Element
Kim, J.L., Nikolov, D.B., Burley, S.K.(1993) Nature 365: 520-527
- PubMed: 8413605
- DOI: https://doi.org/10.1038/365520a0
- Primary Citation of Related Structures:
1VTL - PubMed Abstract:
The three-dimensional structure of a TATA-box binding polypeptide complexed with the TATA element of the adenovirus major late promoter has been determined by X-ray crystallography at 2.25 A resolution. Binding of the saddle-shaped protein induces a conformational change in the DNA, inducing sharp kinks at either end of the sequence TATAAAAG. Between the kinks, the right-handed double helix is smoothly curved and partially unwound, presenting a widened minor groove to TBP's concave, antiparallel beta-sheet. Side-chain/base interactions are restricted to the minor groove, and include hydrogen bonds, van der Waals contacts and phenylalanine-base stacking interactions.
- Laboratory of Molecular Biophysics, Rockefeller University, New York, New York 10021.
Organizational Affiliation: