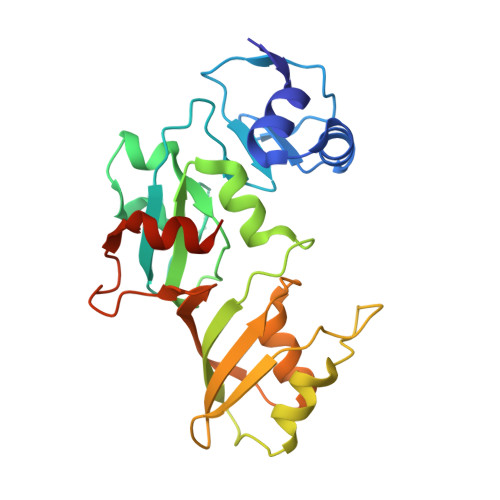
Structure of the pseudouridine synthase RsuA from Haemophilus influenzae.
Matte, A., Louie, G.V., Sivaraman, J., Cygler, M., Burley, S.K.(2005) Acta Crystallogr Sect F Struct Biol Cryst Commun 61: 350-354
- PubMed: 16511038
- DOI: https://doi.org/10.1107/S1744309105005920
- Primary Citation of Related Structures:
1VIO - PubMed Abstract:
The structure of the pseudouridine synthase RsuA from Haemophilus influenza, which catalyzes the conversion of uridine to pseudouridine at a single position within 16S ribosomal RNA, has been determined at 1.59 A resolution and compared with that of Escherichia coli RsuA. The H. influenza enzyme contains an N-terminal S4-like alpha3beta4 domain followed by a catalytic domain, as observed in the structure of E. coli RsuA. Whereas the individual domains of E. coli and H. influenza RsuA are structurally similar, their relative spatial disposition differs greatly between the two structures. The former displays an extended open conformation with no direct contacts between the domains, while the latter is in a closed conformation with a large interface between the two domains. Domain closure presents several basic and polar residues into a putative RNA-binding cleft. It is proposed that this relative repositioning of the S4 and catalytic domains is used to modulate the shape and size of the rRNA-binding site in RsuA and in other pseudouridine synthases possessing S4 domains.
- Biotechnology Research Institute, Montreal, Quebec H4P 2R2, Canada.
Organizational Affiliation: