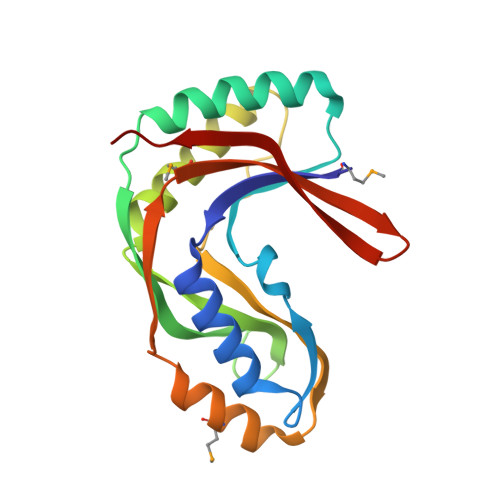
The structure of Pyrococcus horikoshii 2'-5' RNA ligase at 1.94 A resolution reveals a possible open form with a wider active-site cleft
Gao, Y.G., Yao, M., Okada, A., Tanaka, I.(2006) Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 1196-1200
- PubMed: 17142895
- DOI: https://doi.org/10.1107/S1744309106046616
- Primary Citation of Related Structures:
1VGJ - PubMed Abstract:
Bacterial and archaeal 2'-5' RNA ligases, members of the 2H phosphoesterase superfamily, catalyze the linkage of the 5' and 3' exons via a 2'-5'-phosphodiester bond during tRNA-precursor splicing. The crystal structure of the 2'-5' RNA ligase PH0099 from Pyrococcus horikoshii OT3 was solved at 1.94 A resolution (PDB code 1vgj). The molecule has a bilobal alpha+beta arrangement with two antiparallel beta-sheets constituting a V-shaped active-site cleft, as found in other members of the 2H phosphoesterase superfamily. The present structure was significantly different from that determined previously at 2.4 A resolution (PDB code 1vdx) in the active-site cleft; the entrance to the cleft is wider and the active site is easily accessible to the substrate (RNA precursor) in our structure. Structural comparison with the 2'-5' RNA ligase from Thermus thermophilus HB8 also revealed differences in the RNA precursor-binding region. The structural differences in the active-site residues (tetrapeptide motifs H-X-T/S-X) between the members of the 2H phosphoesterase superfamily are discussed.
- Faculty of Advanced Life Sciences, Graduate School of Life Sciences, Hokkaido University, Sapporo 060-0810, Japan.
Organizational Affiliation: