Structural Basis for Thermostability of Pectate Lyase from Bacillus sp. TS 47
Nakaniwa, T., Tada, T., Yamaguchi, A., Kitatani, T., Takao, M., Sakai, T., Nishimura, K.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| pectate lyase 47 | 416 | Bacillus sp. TS-47 | Mutation(s): 0 EC: 4.2.2.2 | 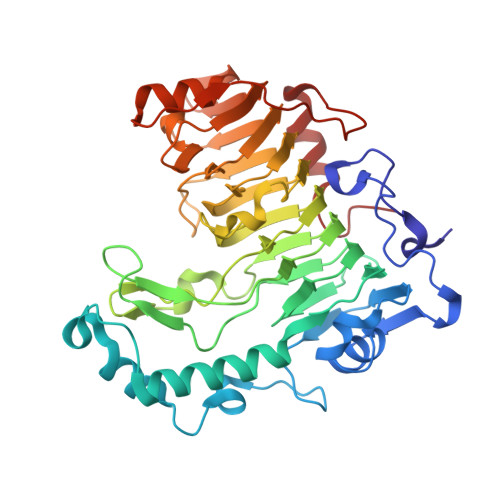 | |
UniProt | |||||
Find proteins for Q9AJM4 (Bacillus sp. TS-47) Explore Q9AJM4 Go to UniProtKB: Q9AJM4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9AJM4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CA Query on CA | B [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.61 | α = 90 |
| b = 58.61 | β = 90 |
| c = 229.42 | γ = 120 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| SCALA | data scaling |
| AMoRE | phasing |
| CNS | refinement |
| CCP4 | data scaling |