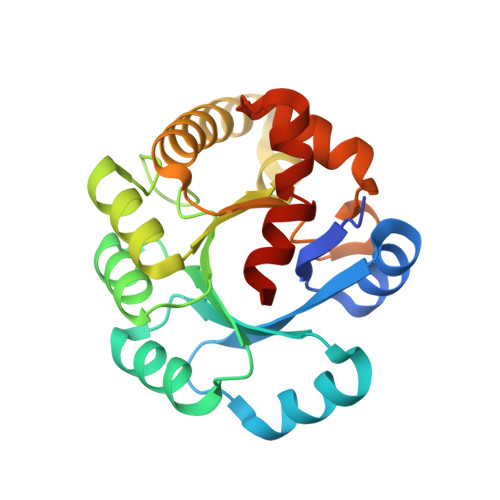
Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3
Takagi, H., Watanabe, M., Kakuta, Y., Kamachi, R., Numata, T., Tanaka, I., Kimura, M.(2004) Biochem Biophys Res Commun 319: 787-794
- PubMed: 15184052
- DOI: https://doi.org/10.1016/j.bbrc.2004.05.055
- Primary Citation of Related Structures:
1V77 - PubMed Abstract:
Ribonuclease P (RNase P) is a ribonucleoprotein complex involved in the processing of pre-tRNA. Protein Ph1877p is one of essential components of the hyperthermophilic archaeon Pyrococcus horikoshii OT3 RNase P [Biochem. Biophys. Res. Commun. 306 (2003) 666]. The crystal structure of Ph1877p was determined at 1.8A by X-ray crystallography and refined to a crystallographic R factor of 22.96% (Rfree of 26.77%). Ph1877p forms a TIM barrel structure, consisting of ten alpha-helices and seven beta-strands, and has the closest similarity to the TIM barrel domain of Escherichia coli cytosine deaminase with a root-mean square deviation of 3.0A. The protein Ph1877p forms an oblate ellipsoid, approximate dimensions being 45Ax43Ax39A, and the electrostatic representation indicated the presence of several clusters of positively charged amino acids present on the molecular surface. We made use of site-directed mutagenesis to assess the role of twelve charged amino acids, Lys42, Arg68, Arg87, Arg90, Asp98, Arg107, His114, Lys123, Lys158, Arg176, Asp180, and Lys196 related to the RNase P activity. Individual mutations of Arg90, Arg107, Lys123, Arg176, and Lys196 by Ala resulted in reconstituted particles with reduced enzymatic activities (32-48%) as compared with that reconstituted RNase P by wild-type Ph1877p. The results presented here provide an initial step for definite understanding of how archaeal and eukaryotic RNase Ps mediate substrate recognition and process 5'-leader sequence of pre-tRNA.
- Laboratory of Biochemistry, Department of Bioscience and Biotechnology, Faculty of Agriculture, Graduate School, Kyushu University, Fukuoka 812-8581, Japan.
Organizational Affiliation: