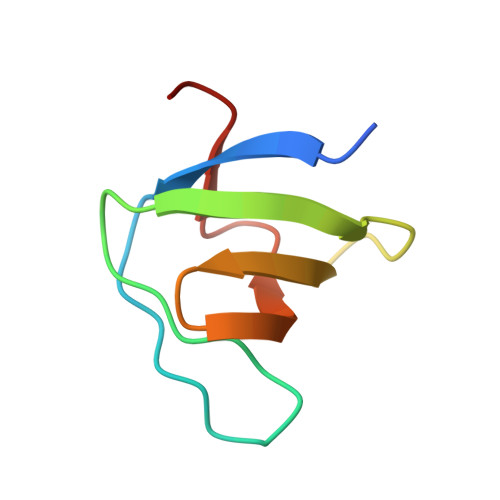
Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain.
Kaneko, T., Kumasaka, T., Ganbe, T., Sato, T., Miyazawa, K., Kitamura, N., Tanaka, N.(2003) J Biological Chem 278: 48162-48168
- PubMed: 13129930
- DOI: https://doi.org/10.1074/jbc.M306677200
- Primary Citation of Related Structures:
1UJ0 - PubMed Abstract:
Although some exceptional motifs have been identified, it is well known that the PXXP motif is the motif of ligand proteins generally recognized by the Src homology 3 (SH3) domain. SH3-ligand interactions are usually weak, with ordinary KD approximately 10 microM. The structural basis for a tight and specific association (KD = 0.24 microm) between Gads SH3 and a novel motif, PX(V/I)(D/N)RXXKP, was revealed in a previous structural analysis of the complex formed between them. In this paper, we report the crystal structure of the signal transducing adaptor molecule-2 (STAM2) SH3 domain in complex with a peptide with a novel motif derived from a ligand protein, UBPY. The derived KD value for this complex is 27 microM. The notable difference in affinity for these parallel complexes may be explained because the STAM2 SH3 structure does not provide a specificity pocket for binding, whereas the Gads SH3 structure does. Instead, the structure of STAM2 SH3 is analogous to that of Grb2 SH3 which, in addition to normal PXXP ligands, has also been shown to moderately recognize the novel motif discussed herein. Thus, the extremely tight interaction observed between Gads SH3 and the novel motif is caused not by an innate ability of the novel motif but rather by an evolutionary change in the Gads SH3 domain. Instead, SH3 domains of STAM2 and Grb2 retain the moderate characteristics of recognizing their ligand proteins like other SH3 domains for appropriate transient interactions between signaling molecules.
- Department of Life Science, Graduate School of Bioscience and Biotechnology, Tokyo Institute of Technology, Yokohama 226-8501, Japan.
Organizational Affiliation: