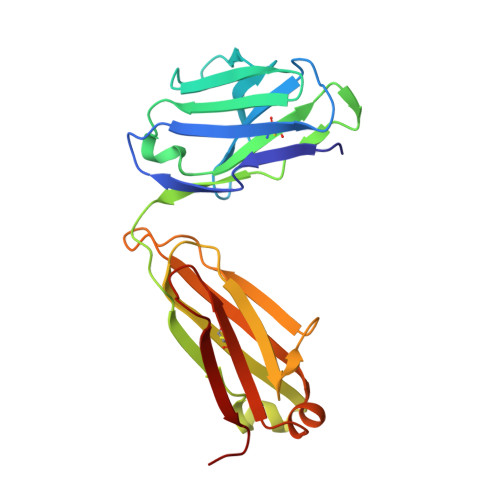
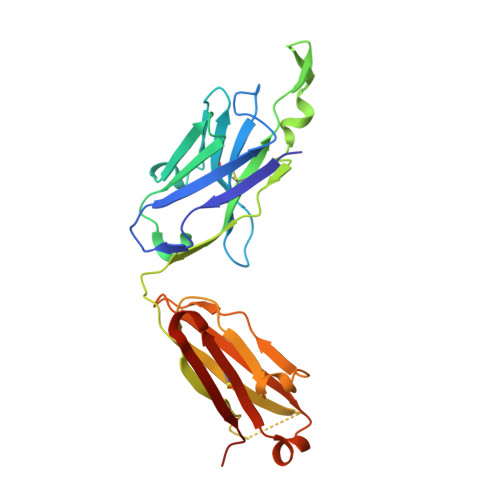
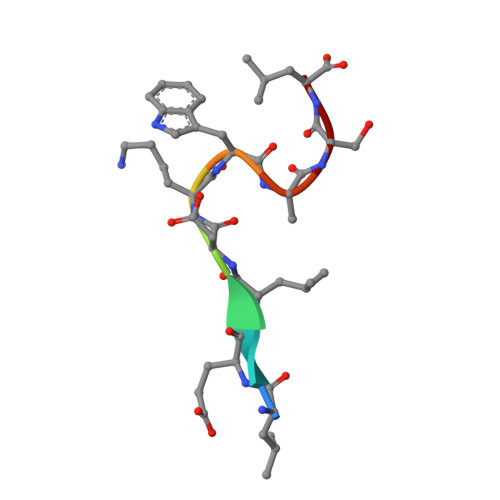
Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptides and Peptide Mimetics
Bryson, S., Hynes, R.C., Cunningham, A., Dong, A., Julien, J.-P., Kunert, R., Katinger, H., Klein, M., Pai, E.F.To be published.