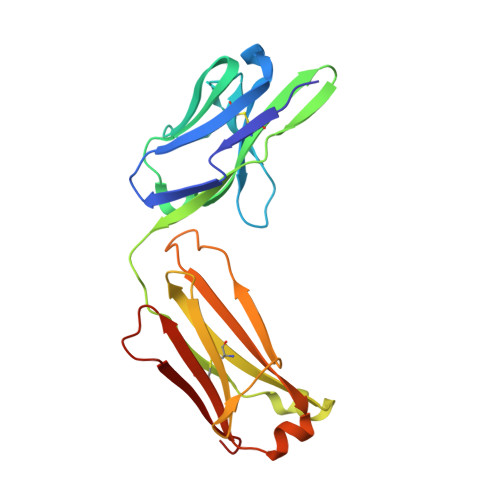
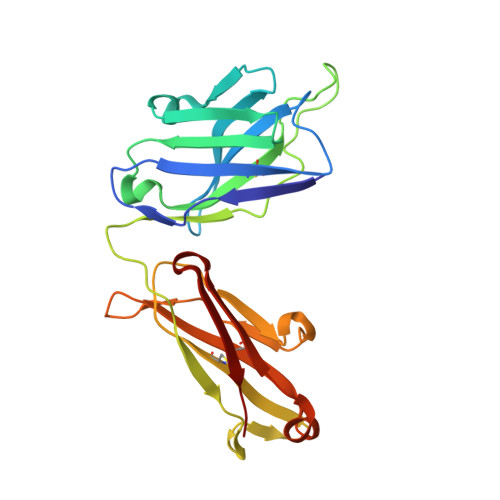
Structure of the Fab fragment of F105, a broadly reactive anti-human immunodeficiency virus (HIV) antibody that recognizes the CD4 binding site of HIV type 1 gp120.
Wilkinson, R.A., Piscitelli, C., Teintze, M., Cavacini, L.A., Posner, M.R., Lawrence, C.M.(2005) J Virol 79: 13060-13069
- PubMed: 16189008
- DOI: https://doi.org/10.1128/JVI.79.20.13060-13069.2005
- Primary Citation of Related Structures:
1U6A - PubMed Abstract:
We have determined the crystal structure of the Fab fragment from F105, a broadly reactive human antibody with limited potency that recognizes the CD4 binding site of gp120. The structure reveals an extended CDR H3 loop with a phenylalanine residue at the apex and shows a striking pattern of serine and tyrosine residues. Modeling the interaction between gp120 and F105 suggests that the phenylalanine may recognize the binding pocket of gp120 used by Phe(43) of CD4 and that numerous tyrosine and serine residues form hydrogen bonds with the main chain atoms of gp120. A comparison of the F105 structure to that of immunoglobulin G1 b12, a much more potent and broadly neutralizing antibody with an overlapping epitope, suggests similarities that contribute to the broad recognition of human immunodeficiency virus by both antibodies. While the putative epitope for F105 shows significant overlap with that predicted for b12, it appears to differ from the b12 epitope in extending across the interface between the inner and outer domains of gp120. In contrast, the CDR loops of b12 appear to interact predominantly with the outer domain of gp120. The difference between the predicted epitopes for b12 and F105 suggests that the unique potency of b12 may arise from its ability to avoid the interface between the inner and outer domains of gp120.
- Department of Chemistry and Biochemistry, Montana State University, Bozeman, 59717, USA.
Organizational Affiliation: