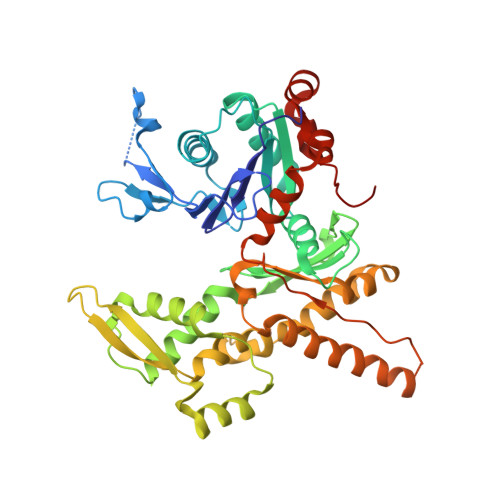
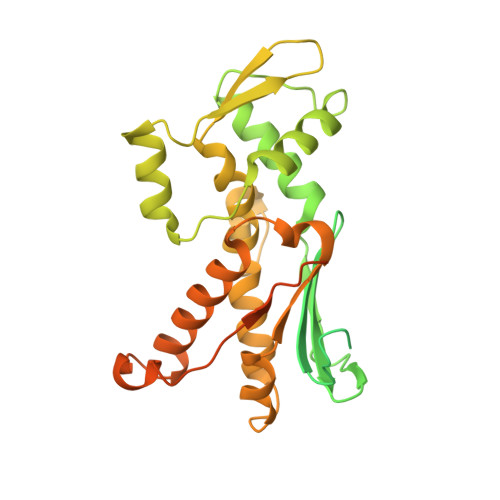
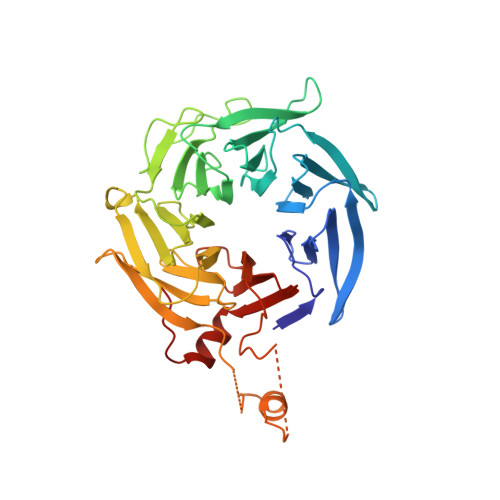
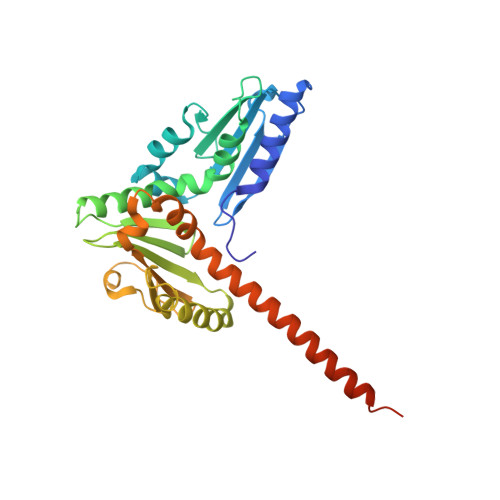
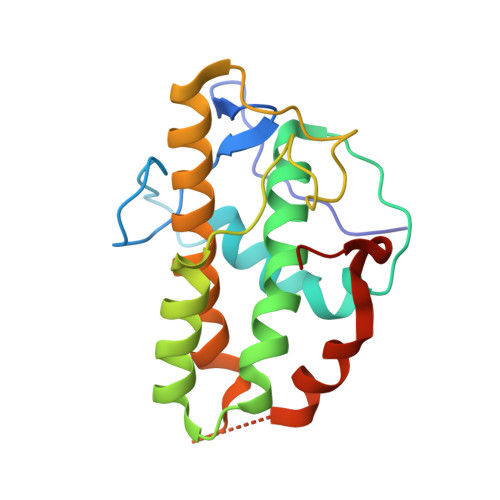
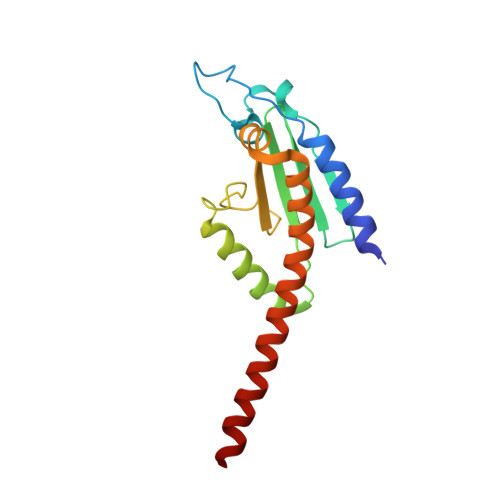
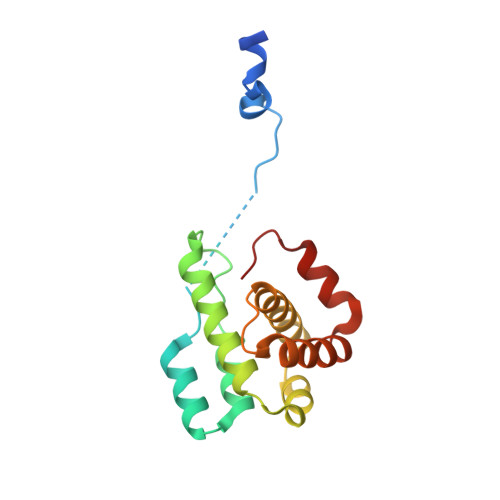
Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP
Nolen, B.J., Littlefield, R.S., Pollard, T.D.(2004) Proc Natl Acad Sci U S A 101: 15627-15632
- PubMed: 15505213
- DOI: https://doi.org/10.1073/pnas.0407149101
- Primary Citation of Related Structures:
1TYQ, 1U2V - PubMed Abstract:
Actin-related protein (Arp) 2/3 complex stimulates formation of actin filaments at the leading edge of motile cells. Nucleation of filaments depends on hydrolysis of ATP bound to Arp2. Here we report crystal structures of Arp2/3 complex with bound ATP or ADP. The nucleotides are immobilized on the face of subdomains 3 and 4 of Arp2, whereas subdomains 1 and 2 are flexible and absent from the electron density maps. This flexibility may explain why Arp2 does not hydrolyze ATP until the complex is activated. ATP stabilizes a relatively closed conformation of Arp3 with the gamma-phosphate bridging loops from opposite sides of the cleft. ADP binds Arp3 in a unique conformation that favors an open cleft, revealing a conformational change that may occur in actin and Arps when ATP is hydrolyzed and phosphate dissociates. These structures provide the an opportunity to compare all nucleotide-binding states in an actin-related protein and give insights into the function of both the Arp2/3 complex and actin.
- Departments of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT 06511, USA.
Organizational Affiliation: