Thiamin Biosynthesis in Bacillus subtilis: Structure of the Thiazole Synthase/Sulfur Carrier Protein Complex
Settembre, E.C., Dorrestein, P.C., Zhai, H., Chatterjee, A., McLafferty, F.W., Begley, T.P., Ealick, S.E.(2004) Biochemistry 43: 11647-11657
- PubMed: 15362849
- DOI: https://doi.org/10.1021/bi0488911
- Primary Citation of Related Structures:
1TYG - PubMed Abstract:
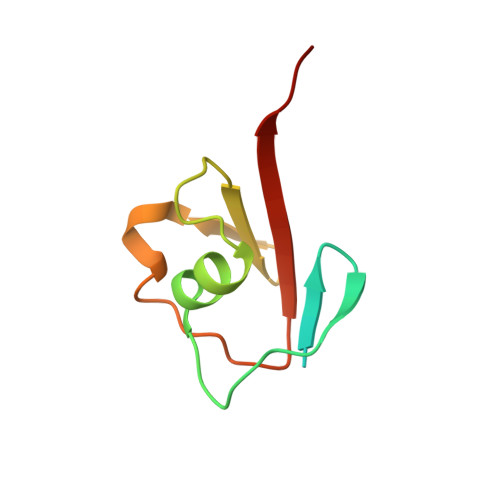
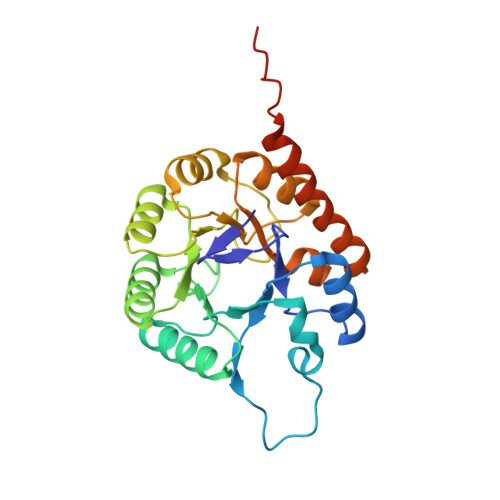
Thiazole synthase is the key enzyme involved in the formation of the thiazole moiety of thiamin pyrophosphate. We have determined the structure of this enzyme in complex with ThiS, the sulfur carrier protein, at 3.15 A resolution. Thiazole synthase is a tetramer with 222 symmetry. The monomer is a (betaalpha)(8) barrel with similarities to the aldolase class 1 and flavin mononucleotide dependent oxidoreductase and phosphate binding superfamilies. The sulfur carrier protein (ThiS) is a compact protein with a fold similar to that of ubiquitin. The structure allowed us to model the substrate, deoxy-D-xylulose 5-phosphate (DXP), in the active site. This model identified Glu98 and Asp182 as new active site residues likely to be involved in the catalysis of thiazole formation. The function of these residues was probed by mutagenesis experiments, which confirmed that both residues are essential for thiazole formation and identified Asp182 as the base involved in the deprotonation at C3 of the thiazole synthase DXP imine. Comparison of the ThiS binding surface to the surface of ubiquitin identified a conserved hydrophobic patch of unknown function on ubiquitin that may be involved in complex formation between ubiquitin and one of its binding partners.
- Department of Chemistry and Chemical Biology, Cornell University, Ithaca, New York 14853, USA.
Organizational Affiliation: