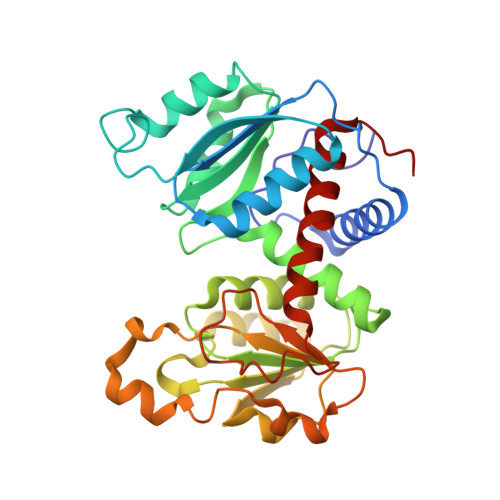
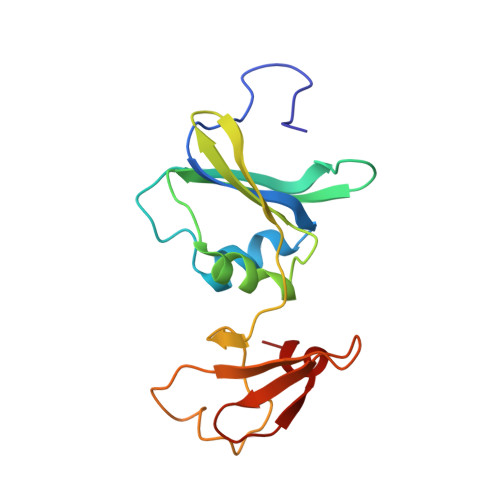
Monitoring the Transition from the T to the R State in E.coli Aspartate Transcarbamoylase by X-ray Crystallography: Crystal Structures of the E50A Mutant Enzyme in Four Distinct Allosteric States.
Stieglitz, K., Stec, B., Baker, D.P., Kantrowitz, E.R.(2004) J Mol Biology 341: 853-868
- PubMed: 15288791
- DOI: https://doi.org/10.1016/j.jmb.2004.06.002
- Primary Citation of Related Structures:
1TTH, 1TU0, 1TUG - PubMed Abstract:
A detailed description of the transition that allosteric enzymes undergo constitutes a major challenge in structural biology. We have succeeded in trapping four distinct allosteric states of a mutant enzyme of Escherichia coli aspartate transcarbomylase and determining their structures by X-ray crystallography. The mutant version of aspartate transcarbamoylase in which Glu50 in the catalytic chains was replaced by Ala destabilizes the native R state and shifts the equilibrium towards the T state. This behavior allowed the use of substrate analogs such as phosphonoacetamide and malonate to trap the enzyme in T-like and R-like structures that are distinct from the T-state structure of the wild-type enzyme (as represented by the structure of the enzyme with CTP bound and the R-state structure as represented by the structure with N-(phosphonacetyl)-L-aspartate bound). These structures shed light on the nature and the order of internal structural rearrangements during the transition from the T to the R state. They also suggest an explanation for diminished activity of the E50A enzyme and for the change in reaction mechanism from ordered to random for this mutant enzyme.
- Department of Chemistry, Merkert Chemistry Center, Boston College, Chestnut Hill, MA 02467, USA.
Organizational Affiliation: