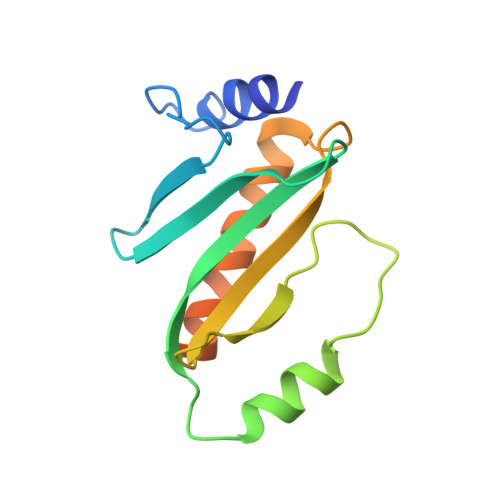
Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2.
Phan, J., Tropea, J.E., Waugh, D.S.(2004) Acta Crystallogr D Biol Crystallogr 60: 1591-1599
- PubMed: 15333930
- DOI: https://doi.org/10.1107/S0907444904017597
- Primary Citation of Related Structures:
1TTW - PubMed Abstract:
Pathogenic Yersinia species use a type III secretion system to inject cytotoxic effector proteins directly into the cytosol of mammalian cells, where they neutralize the innate immune response by interfering with the signal-transduction pathways that control phagocytosis and inflammation. To be exported efficiently, some effectors must transiently associate with cognate cytoplasmic secretion chaperones. SycH is the chaperone for YopH, a potent eukaryotic-like protein tyrosine phosphatase that is essential for virulence. SycH also binds two negative regulators of type III secretion, YscM1 and YscM2, both of which share significant sequence homology with the chaperone-binding domain of YopH. Here, the structure of a complex between SycH and a stable fragment of YscM2 that was designed on the basis of limited proteolysis experiments is presented. The overall fold of SycH is very similar to the structures of other homodimeric secretion chaperones that have been determined to date. YscM2 wraps around SycH in an extended fashion, with some secondary but no tertiary structure, assuming a conformation distinct from the globular fold that it is predicted to adopt in the absence of SycH.
- Protein Engineering Section, Macromolecular Crystallography Laboratory, Center for Cancer Research, National Cancer Institute at Frederick, PO Box B, Frederick, MD 21702, USA.
Organizational Affiliation: