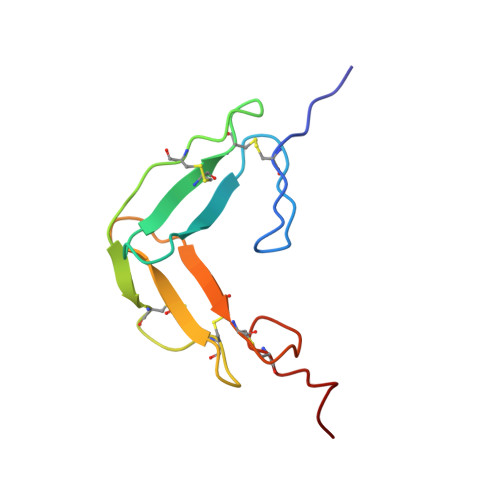
The solution structure and backbone dynamics of the fibronectin type I and epidermal growth factor-like pair of modules of tissue-type plasminogen activator.
Smith, B.O., Downing, A.K., Driscoll, P.C., Dudgeon, T.J., Campbell, I.D.(1995) Structure 3: 823-833
- PubMed: 7582899
- DOI: https://doi.org/10.1016/s0969-2126(01)00217-9
- Primary Citation of Related Structures:
1TPG - PubMed Abstract:
The thrombolytic serine protease tissue-type plasminogen activator (t-PA) is a classical modular protein consisting of three types of domain in addition to the serine protease domain: F1 (homologous to fibronectin type I); G (epidermal growth factor-like) and kringle. Biochemical data suggest that the F1 and G modules play a major role in the binding of t-PA to fibrin and to receptors on hepatocytes. We have derived the solution structure of the F1 and G pair of modules from t-PA by two- and three-dimensional NMR techniques, in combination with dynamical simulated annealing calculations. We have also obtained information about the molecule's backbone dynamics through measurement of amide 15N relaxation parameters. Although the F1 and G modules each adopt their expected tertiary structure, the modules interact intimately to bury a hydrophobic core, and the inter-module linker makes up the third strand of the G module's major beta-sheet. The new structural results allow the interpretation of earlier mutational data relevant to fibrin-binding and hepatocyte-receptor binding.
- Oxford Centre for Molecular Sciences, UK.
Organizational Affiliation: