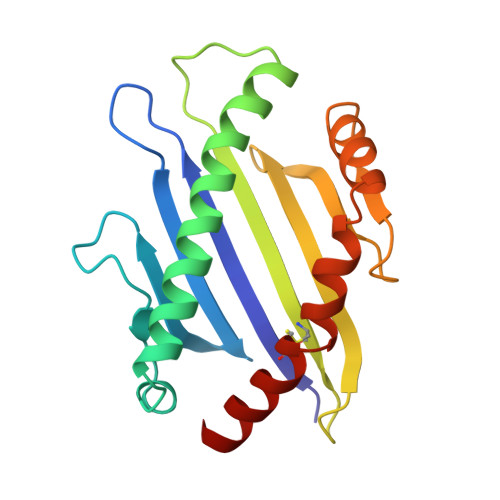
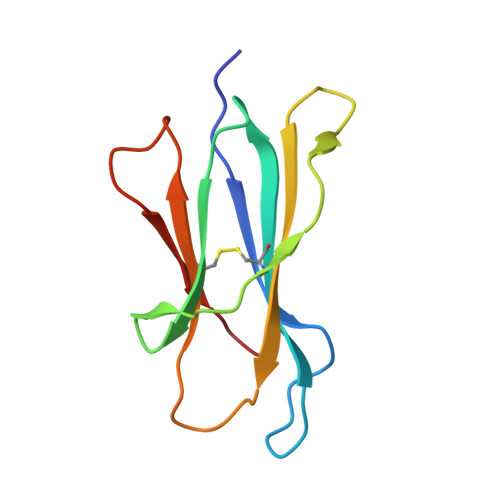
The three-dimensional structure of a class I major histocompatibility complex molecule missing the alpha 3 domain of the heavy chain.
Collins, E.J., Garboczi, D.N., Karpusas, M.N., Wiley, D.C.(1995) Proc Natl Acad Sci U S A 92: 1218-1221
- PubMed: 7862664
- DOI: https://doi.org/10.1073/pnas.92.4.1218
- Primary Citation of Related Structures:
1TMC - PubMed Abstract:
Class I major histocompatibility complex (MHC) molecules are ternary complexes of the soluble serum protein beta 2-microglobulin, MHC heavy chain, and bound peptide. The first two domains (alpha 1, alpha 2) of the heavy chain create the peptide binding cleft and the surface that contacts the T-cell receptor. The third domain (alpha 3) associates with the T-cell co-receptor, CD8, during T-cell recognition. Here we describe the x-ray crystal structure of a human class I MHC molecule, HLA-Aw68, from which the alpha 3 domain has been proteolytically removed. The resulting molecule shows no gross morphological changes compared to the intact protein. A decameric peptide complexed with the intact HLA-Aw68 is seen to bind to the proteolized molecule in the conventional manner, demonstrating that the alpha 3 domain is not required for the structural integrity of the molecule or for peptide binding.
- Department of Molecular and Cellular Biology, Howard Hughes Medical Institute, Harvard University, Cambridge, MA 02138.
Organizational Affiliation: