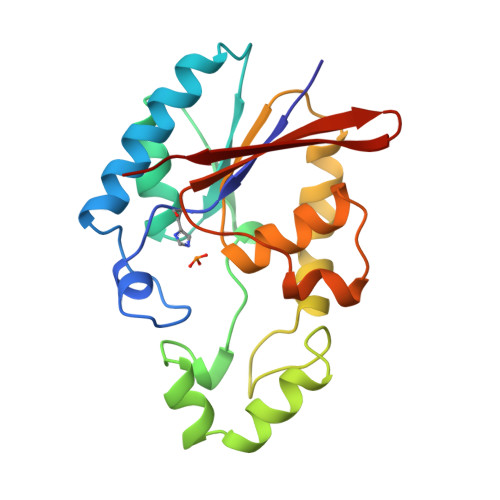
Crystal structure of a trapped phosphoenzyme during a catalytic reaction.
Lee, Y.H., Olson, T.W., Ogata, C.M., Levitt, D.G., Banaszak, L.J., Lange, A.J.(1997) Nat Struct Biol 4: 615-618
- PubMed: 9253407
- DOI: https://doi.org/10.1038/nsb0897-615
- Primary Citation of Related Structures:
1TIP - PubMed Abstract:
The crystal structure of the fructose-2,6-bisphosphatase domain trapped during the reaction reveal a phosphorylated His 258, and a water molecule immobilized by the product, fructose-6-phosphate. The geometry suggests that the dephosphorylation step requires prior removal of the product for an 'associative in-line' phosphoryl transfer to the catalytic water.