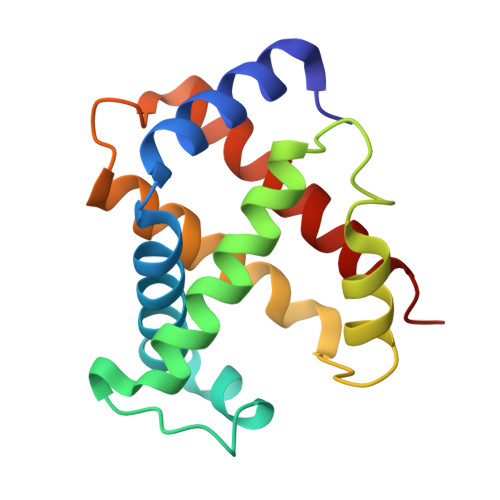
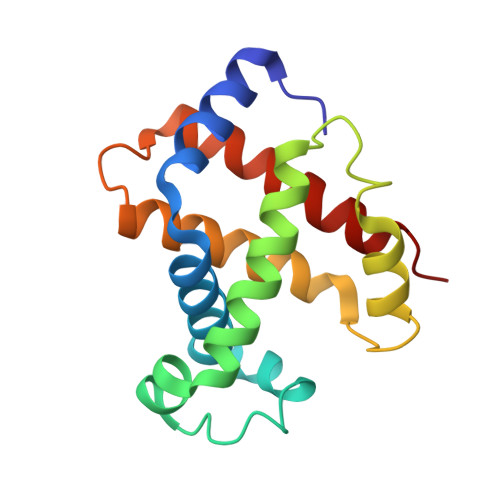
Refinement of a partially oxygenated T state human haemoglobin at 1.5 A resolution.
Waller, D.A., Liddington, R.C.(1990) Acta Crystallogr B 46: 409-418
- PubMed: 2383372
- DOI: https://doi.org/10.1107/s0108768190000313
- Primary Citation of Related Structures:
1THB - PubMed Abstract:
The degree of ligation of T state human haemoglobin crystals is reduced by inositol hexaphosphate (IHP). The structure of a partially ligated haemoglobin has been refined using fast Fourier restrained-least-squares techniques. Manual interventions were required to escape from local minima and introduce a large number of solvent molecules. Individual isotropic temperature factors were refined for all atoms and the final average atomic temperature factor is 32.3 A2. The final R factor is 19.6% for all data between 10 and 1.5 A. The final model consists of 4560 protein atoms and 313 solvent molecules. The occupancies of the ligand atoms and the anisotropic behaviour of the iron atoms have been refined, demonstrating that the alpha haem groups are only partially ligated and that there is no ligation of the beta haems. Density for the IHP indicates that it is not well ordered even though changes in the ligation and structure of the haemoglobin indicate its presence.
- Department of Chemistry, University of York, Heslington, England.
Organizational Affiliation: