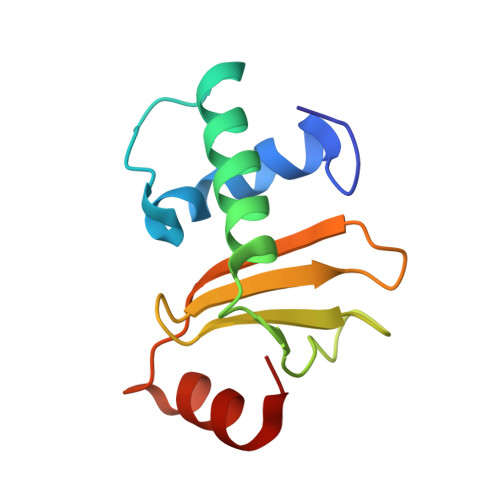
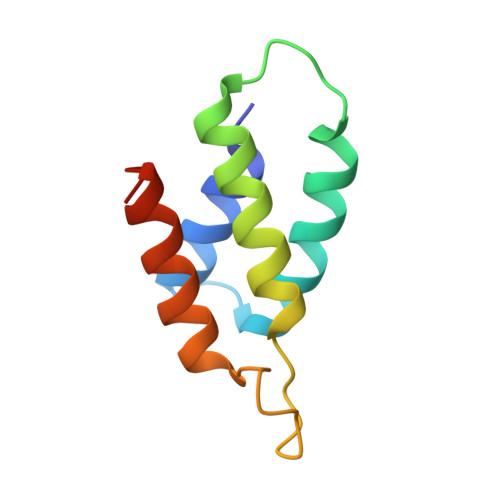
Relation between tRNase activity and the structure of colicin D according to X-ray crystallography
Yajima, S., Nakanishi, K., Takahashi, K., Ogawa, T., Hidaka, M., Kezuka, Y., Nonaka, T., Ohsawa, K., Masaki, H.(2004) Biochem Biophys Res Commun 322: 966-973
- PubMed: 15336558
- DOI: https://doi.org/10.1016/j.bbrc.2004.07.206
- Primary Citation of Related Structures:
1TFK, 1TFO - PubMed Abstract:
Colicin D is a plasmid-encoded proteinaceous toxin which kills sensitive Escherichia coli. Toxicity stems from ribonuclease activity that targets exclusively four isoacceptors of tRNA(Arg) with a cleavage position between 38 and 39 of the corresponding anticodons. Since no other tRNAs with the same sequences at 38 and 39 as tRNA(Arg)s are cleaved, colicin D should be capable of recognizing some higher order structure of tRNAs. We report here two crystal structures of catalytic domains of colicin D which have different N-terminal lengths, both complexed with its cognate inhibitor protein, ImmD. A row of positive charge patches is found on the surface of the catalytic domain, suggestive of the binding site of the tRNAs. This finding, together with our refined tRNase activity experiments, indicates that the catalytic domain starting at position 595 has activity almost equivalent to that of colicin D.
- Department of Bioscience, Tokyo University of Agriculture, Setagaya-ku, Tokyo 156-8502, Japan. yshun@nodai.ac.jp
Organizational Affiliation: