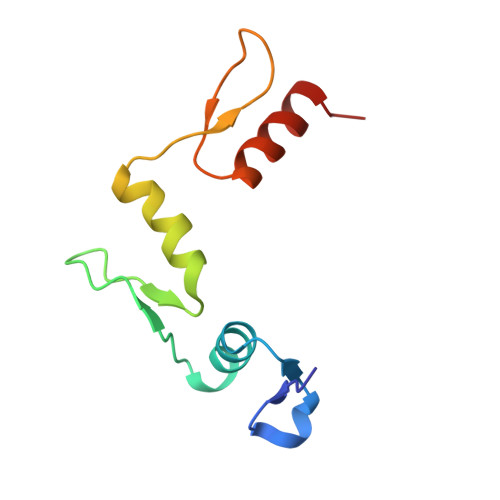
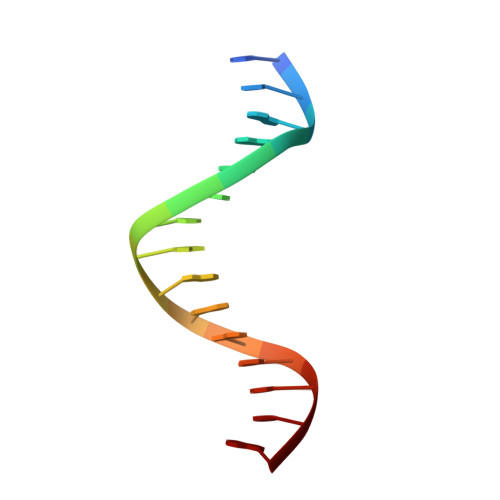
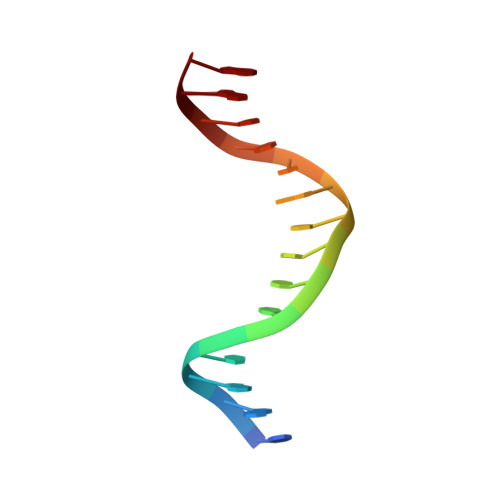
Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Foster, M.P., Wuttke, D.S., Radhakrishnan, I., Case, D.A., Gottesfeld, J.M., Wright, P.E.(1997) Nat Struct Biol 4: 605-608
- PubMed: 9253405
- DOI: https://doi.org/10.1038/nsb0897-605
- Primary Citation of Related Structures:
1TF3 - PubMed Abstract:
The three N-terminal zinc fingers of transcription factor IIIA bind in the DNA major groove. Substantial packing interfaces are formed between adjacent fingers, the linkers lose their intrinsic flexibility upon DNA binding, and several lysine side chains implicated in DNA recognition are dynamically disordered.